Search Count: 23
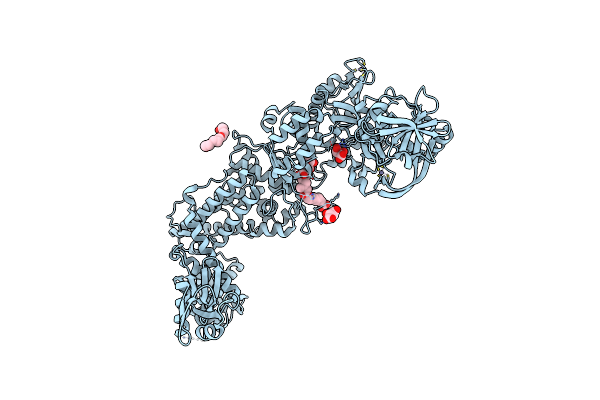 |
Priestia Megaterium Mupirocin-Resistant Isoleucyl-Trna Synthetase 2 Complexed With Mupirocin
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-23 Classification: RNA BINDING PROTEIN Ligands: ZN, MRC, TLA, 1PE |
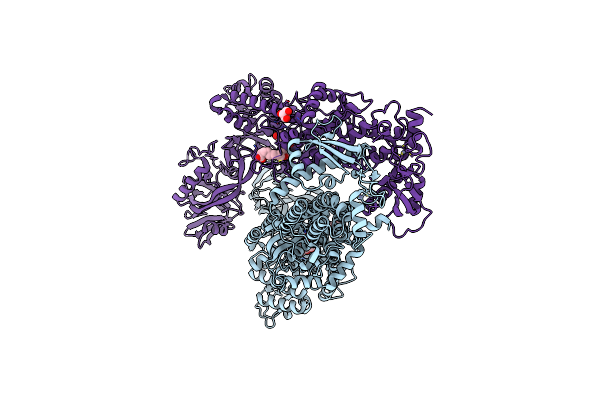 |
Priestia Megaterium Mupirocin-Sensitive Isoleucyl-Trna Synthetase 1 Complexed With Mupirocin
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-08-23 Classification: RNA BINDING PROTEIN Ligands: ZN, MRC, CL, NA, GOL |
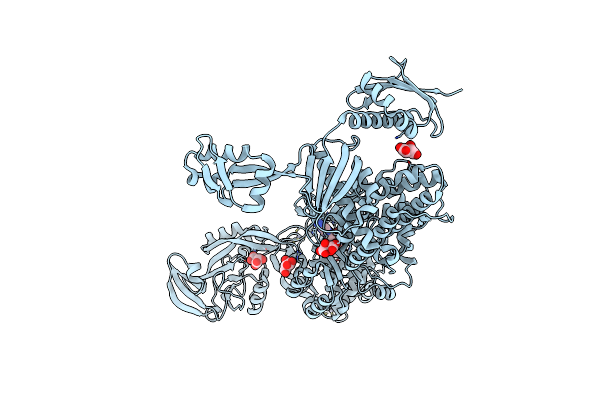
 |
Priestia Megaterium Mupirocin-Resistant Isoleucyl-Trna Synthetase 2 With A Fully-Resolved C-Terminal Trna-Binding Domain Complexed With An Isoleucyl-Adenylate Analogue
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-16 Classification: RNA BINDING PROTEIN Ligands: ZN, ILA, TAR |
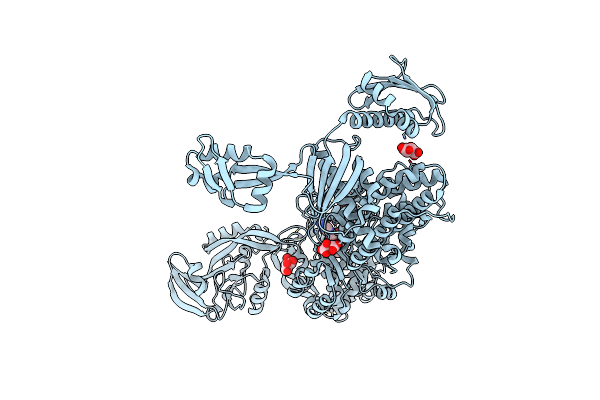
 |
Priestia Megaterium Mupirocin Hyper-Resistant High Motif Mutant Of Type 2 Isoleucyl-Trna Synthetase Complexed With An Isoleucyl-Adenylate Analogue
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-08-16 Classification: RNA BINDING PROTEIN Ligands: ILA, TAR, ZN |
 |
Priestia Megaterium W130Q Mutant Of Type 2 Isoleucyl-Trna Synthetase Complexed With An Isoleucyl-Adenylate Analogue
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-16 Classification: RNA BINDING PROTEIN Ligands: ZN, ILA, TAR |
 |
Priestia Megaterium Mupirocin-Sensitive Isoleucyl-Trna Synthetase 1 Complexed With An Isoleucyl-Adenylate Analogue
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-08-16 Classification: RNA BINDING PROTEIN Ligands: ZN, ILA, SO4, CL, LI, NA |
 |
Priestia Megaterium Inactive High Motif Mutant Of Type1 Isoleucyl-Trna Synthetase Complexed With An Isoleucyl-Adenylate Analogue.
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-08-16 Classification: RNA BINDING PROTEIN Ligands: ZN, ILA, SO4, CL, LI, NA |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs L550A Mutant In Complex With Atp And L-Leucinol
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-09-28 Classification: LIGASE Ligands: EDO, ATP, MG, DCL, ZN |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs L550A Mutant In Complex With The Reaction Intermediate Leu-Amp
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: USB, ZN, MG |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs E169G Mutant In Complex With The Reaction Intermediate Leu-Amp
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, ATP, USB, ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs E169G Mutant In Complex With Atp And L-Leucinol
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, DCL, ATP, ZN, MG |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: ZN, EDO, MG |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: LEU, ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs In Complex With Atp In Conformation 1
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, ATP, ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs In Complex With Atp In Conformation 2
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, ATP, ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs In Complex With The Reaction Intermediate Leu-Amp
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, USB, ATP, ZN, MG |
 |
Crystal Structure Of Neisseria Gonorrhoeae Leurs In Complex With Atp And L-Leucinol
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: EDO, ATP, MG, DCL, ZN |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2022-08-31 Classification: LIGASE Ligands: ZN, MG |

