Search Count: 59
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With N-Pnt-Dnm 15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-09-13 Classification: HYDROLASE Ligands: CL, GOL, PGE, EDO, U4X, SO4 |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Covalent Complex With Tamra Tagged 1,6-Epi-Cylcophellitol Aziridine Activity Based Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-13 Classification: HYDROLASE Ligands: CL, EDO, GOL, U54, NAG, SO4, PGE |
 |
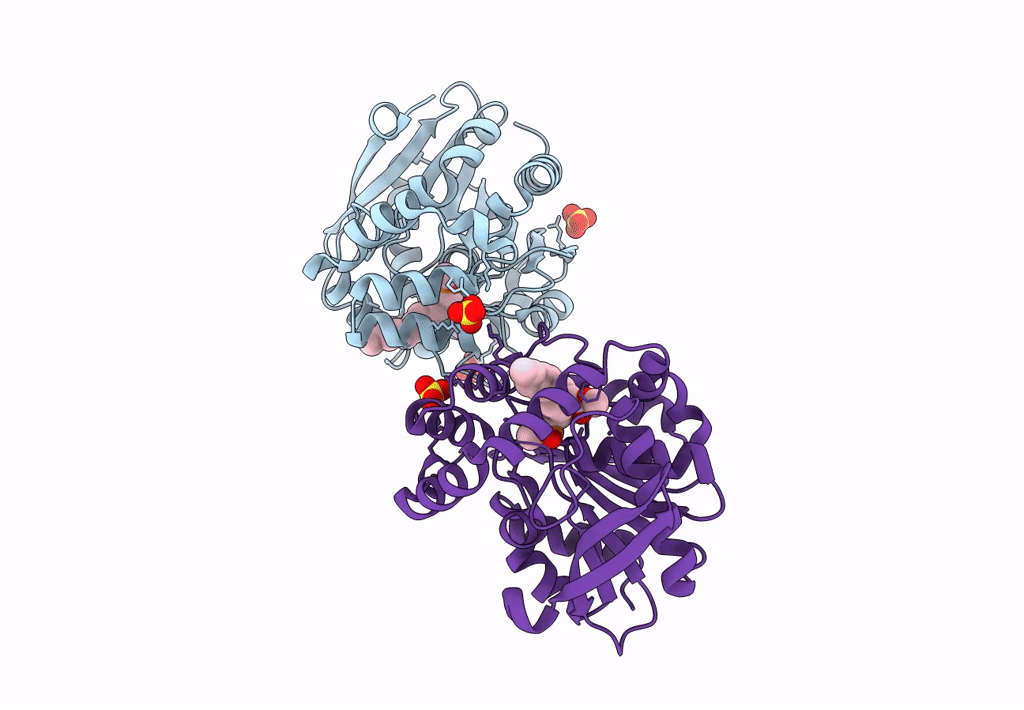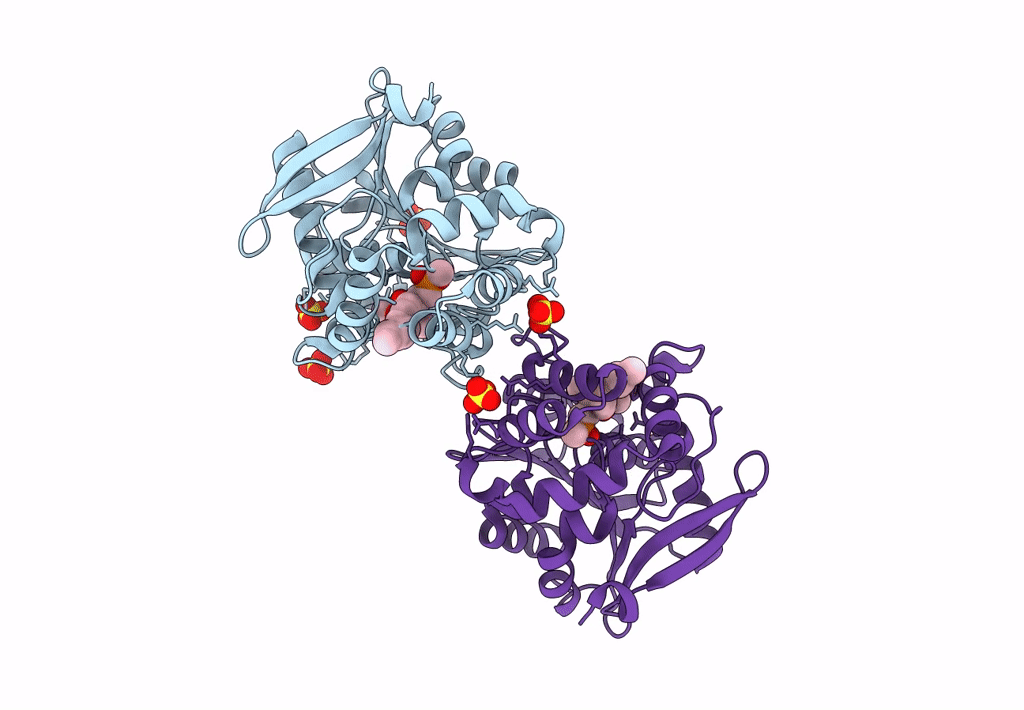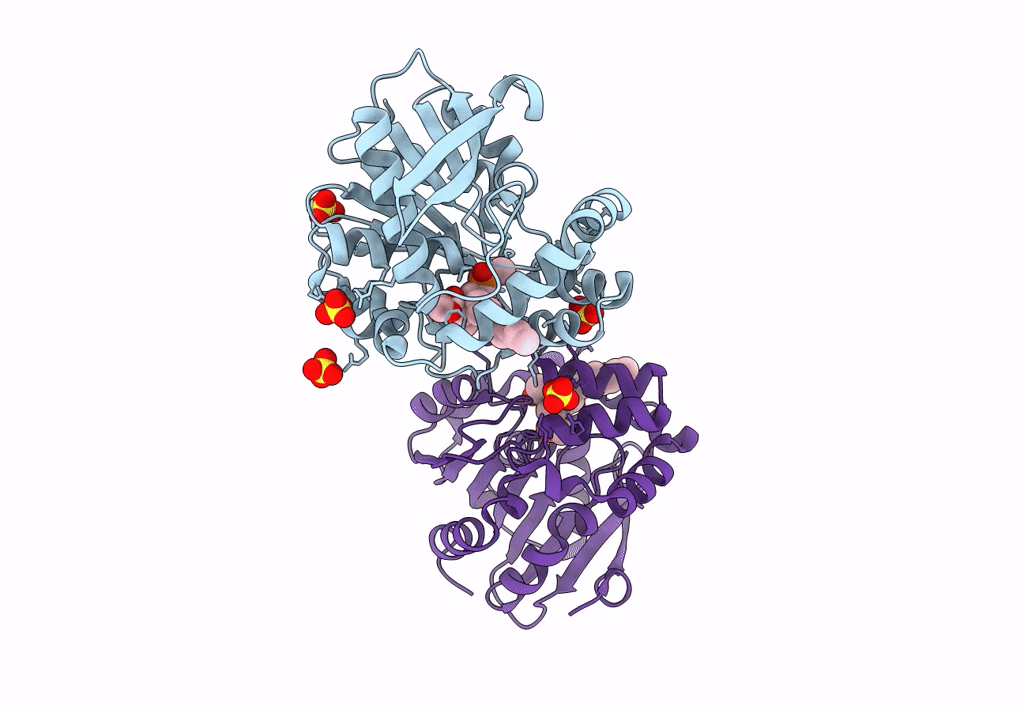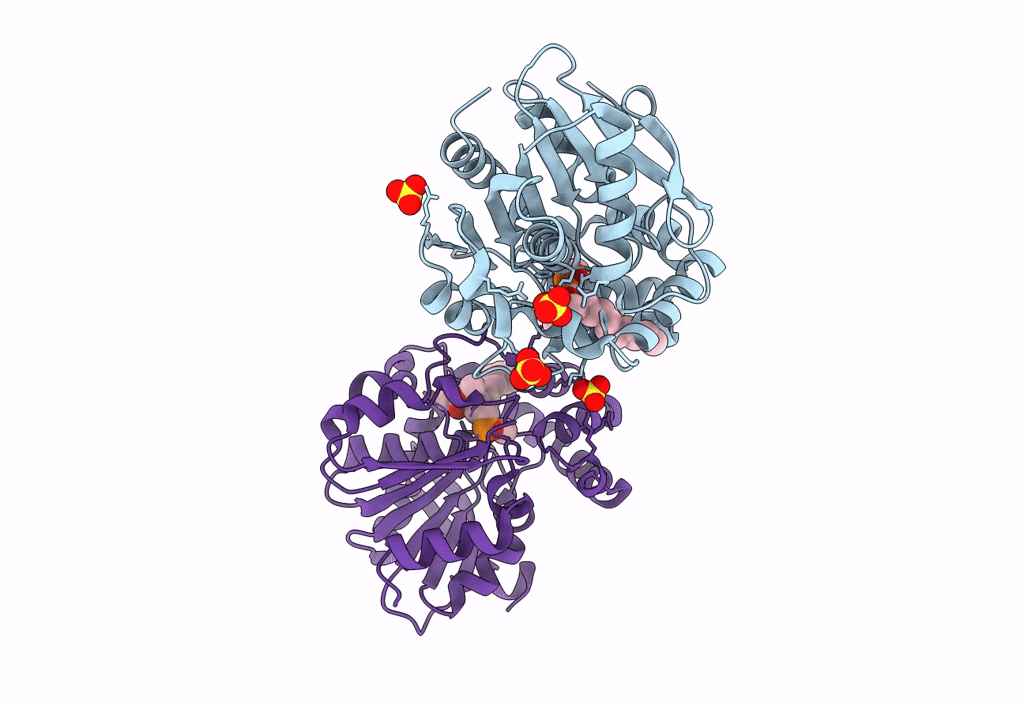
Crystal Structure Of Hsad From Mycobacterium Tuberculosis At 1.96 A Resolution
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: EDO, CL |
 |
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclophostin-Like Inhibitor Cyc7B
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: IY8, SO4 |
 |
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclophostin-Like Inhibitor Cyc8B
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: 9SW, SO4 |
 |
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclipostin-Like Inhibitor Cyc17
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: E9H, SO4 |
 |
Crystal Structure Of Hsad From Mycobacterium Tuberculosis In Complex With Cyclipostin-Like Inhibitor Cyc31
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: IYB, SO4 |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase In Glycoside Hydrolase Family 31, In Complex With Noncovalent Cyclophellitol Sulfamidate Probe Kk131
Organism: Cellvibrio japonicus (strain ueda107)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-07-27 Classification: HYDROLASE Ligands: EDO, OXL, 5OV, SO4 |
 |
Crystal Structure Of Agd31B, Alpha-Transglucosylase In Glycoside Hydrolase Family 31, In Complex With Covalent Cyclophellitol Sulfamidate Probe Kk130
Organism: Cellvibrio japonicus ueda107
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-27 Classification: HYDROLASE Ligands: EDO, PG4, OXL, 56I, SO4 |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With Cyclosulfamidate 4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: SO4, CL, GOL, EDO, YTW, PGE |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With Cyclosulfamidate 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: NAG, 56I, SO4, CL, GOL, EDO, PGE |
 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Bodipy Tagged Cyclophellitol Activity Based Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: SO4, UUH, NAG, EDO, UUE |
 |
Organism: Sudan virus - boniface, sudan,1976, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-02-17 Classification: TRANSFERASE Ligands: GOL, PEG, NA, CL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: NAG, ACT, SO4, CA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: NAG, GOL, ACT, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-04-03 Classification: ISOMERASE Ligands: CA, NAG, GOL, MES, P6G |
 |
Structure Of Maf Glycosyltransferase From Magnetospirillum Magneticum Amb-1
Organism: Magnetospirillum magneticum amb-1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-15 Classification: TRANSFERASE Ligands: SO4, CL, PGE, PG4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: SO4, CL, PGE, PEG, EDO |
 |
Crystal Structure Of Human Lysosomal Acid-Alpha-Glucosidase, Gaa, In Complex With N-Acetyl-Cysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-10-25 Classification: HYDROLASE Ligands: SC2, SO4, CL, PGE, GOL, EDO |

