Search Count: 62
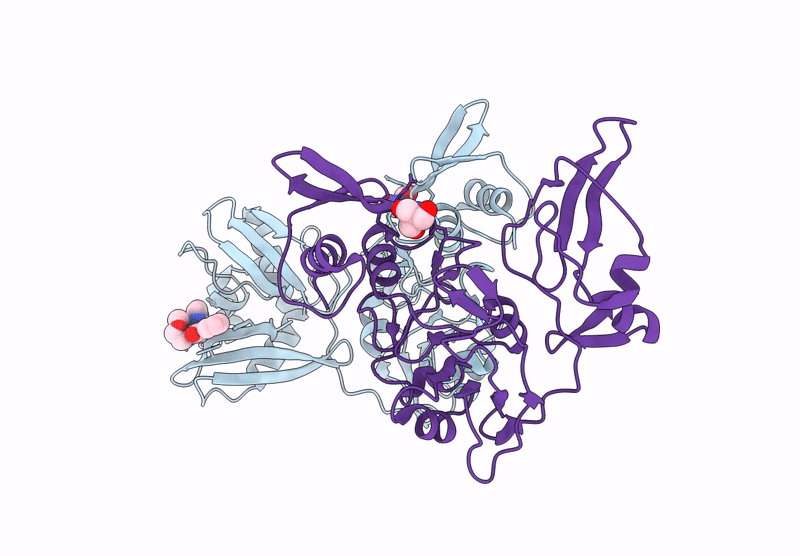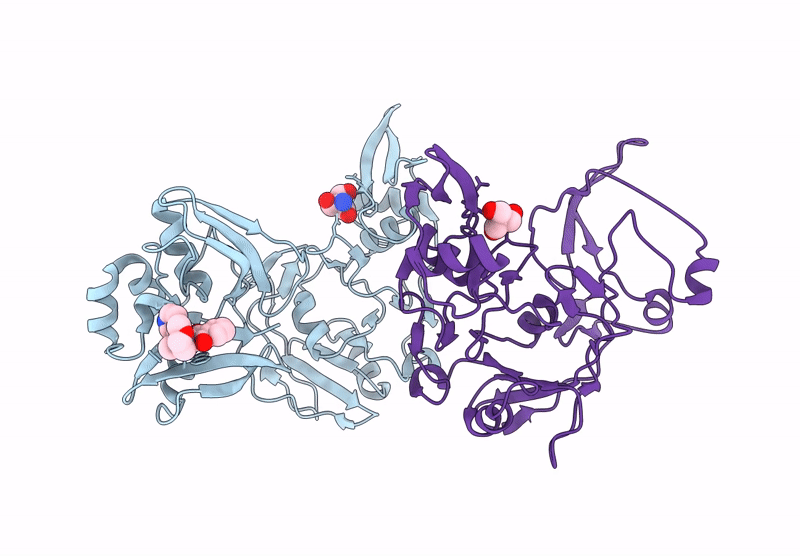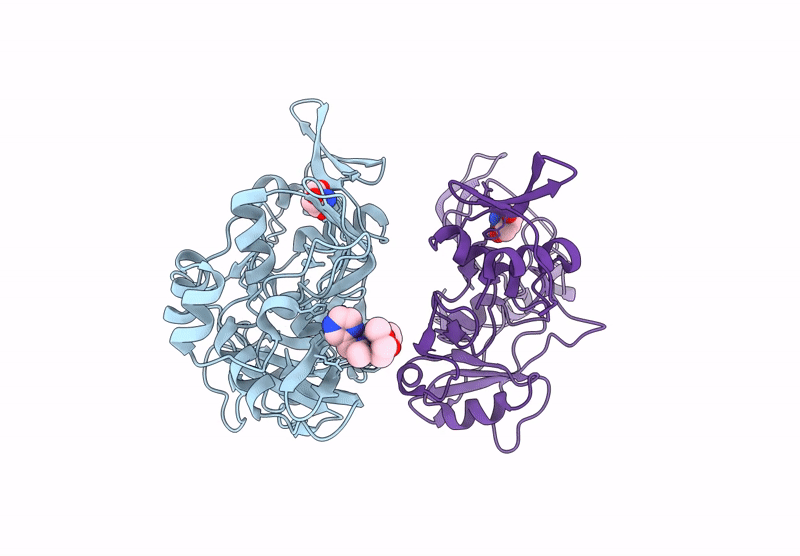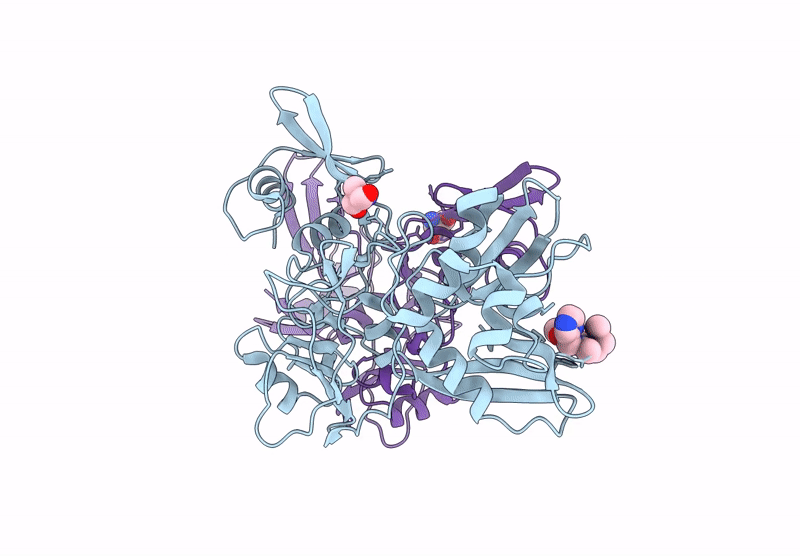 |
Crystal Structure Of Nsp15 Endoribonuclease From Sars Cov-2 In Complex With Sepantronium (Ym-155)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: TRS, GXU |
 |
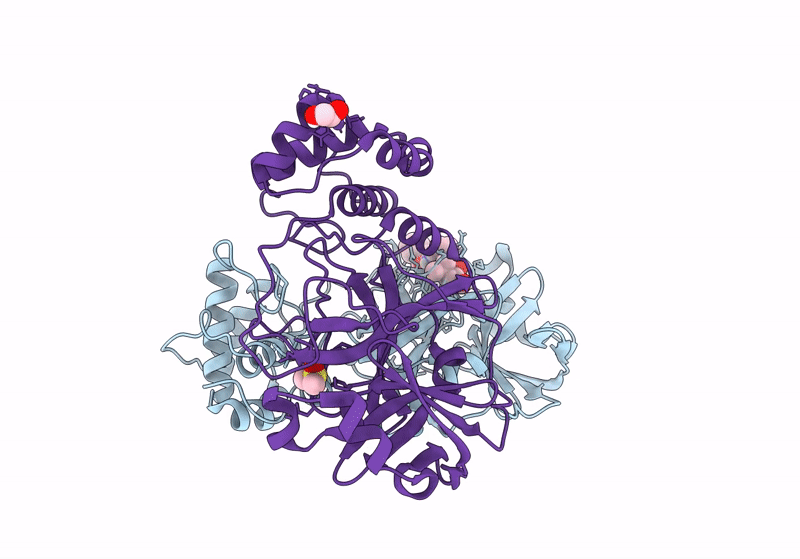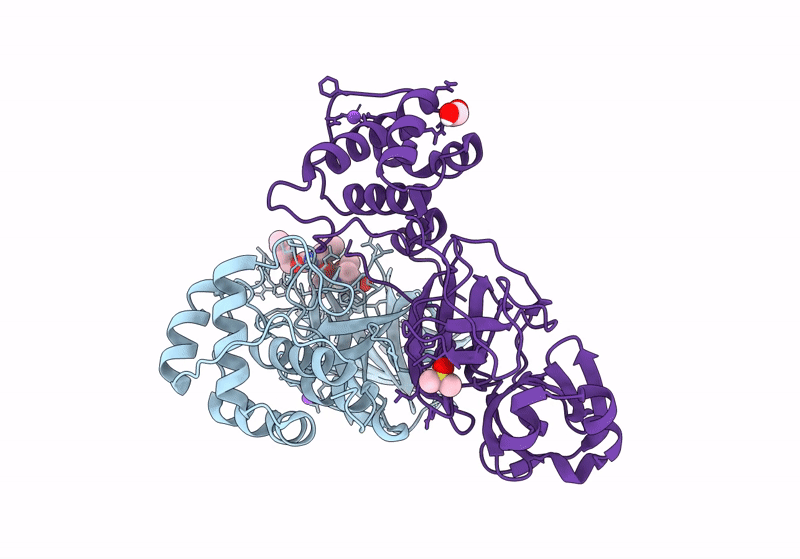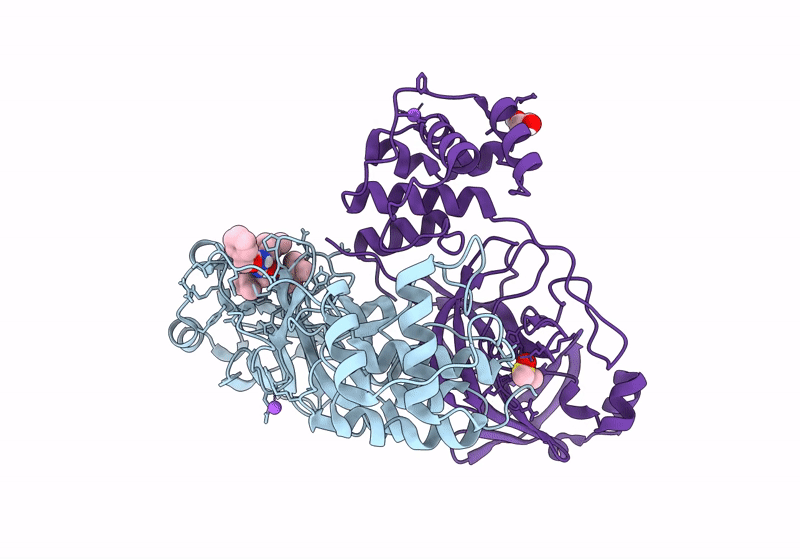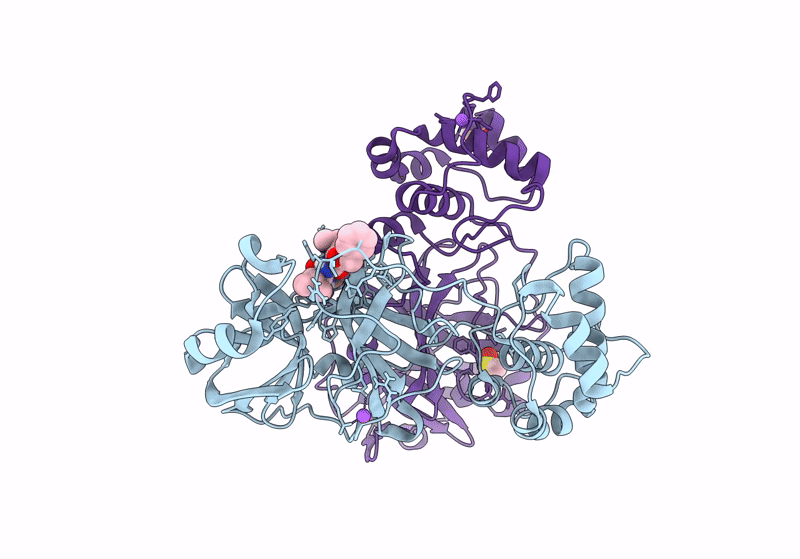
Crystal Structure Of Nsp15 Endoribonuclease From Sars Cov-2 In Complex With Tas-103
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:2.00 Å Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: A1IUV, PO4, EDO |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: CL, X77, EDO, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: DMS, EDO, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: 9M5, EDO, ACT, DMS, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm Mg-132, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm X77, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ACT, EDO, CL, X77, NA |
 |
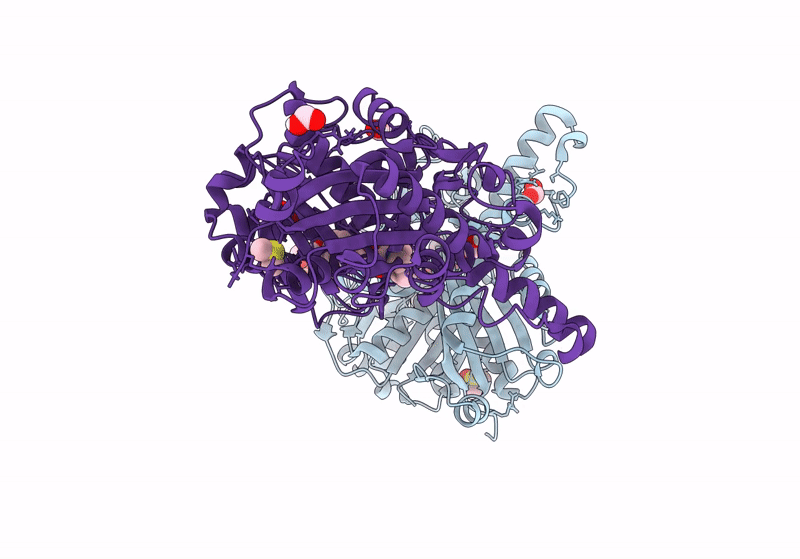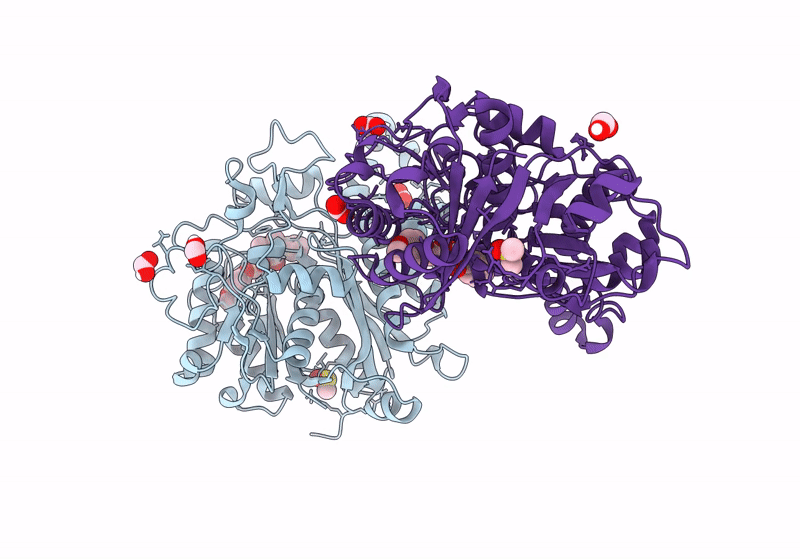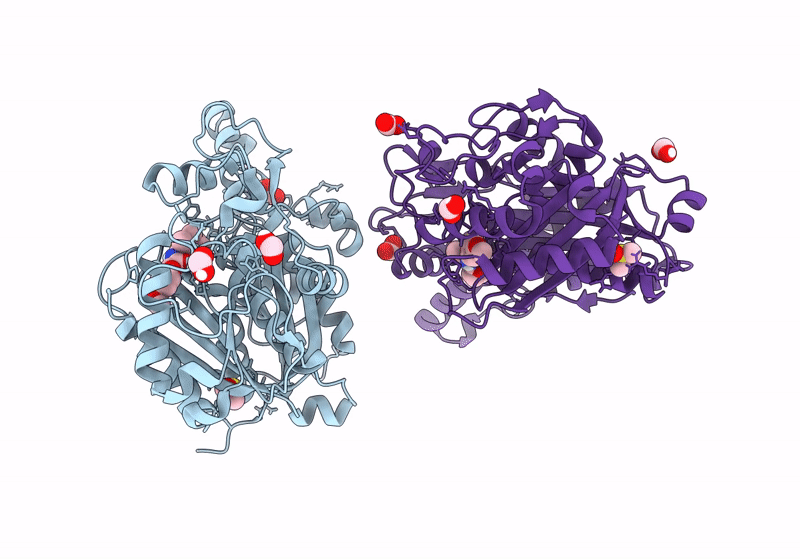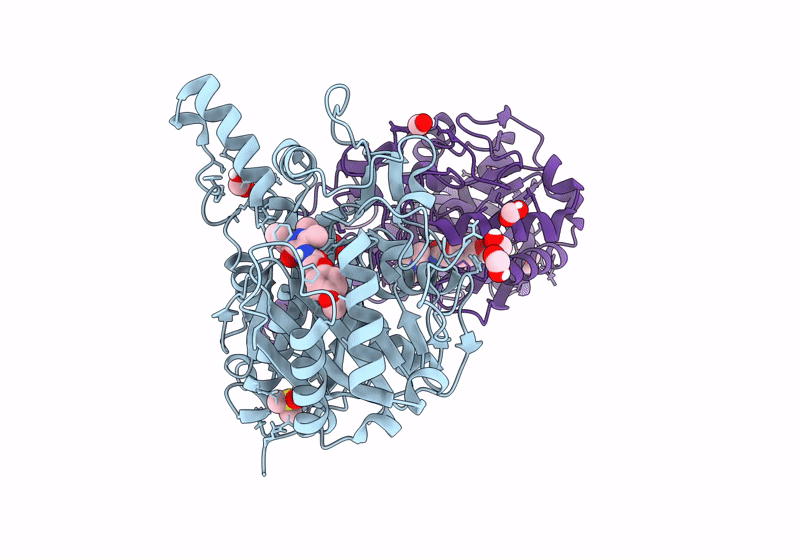
Pseudomonas Aeruginosa Fabf C164A In Complex With 3-Amino-N-(1,5-Dimethyl-3-Oxo-2-Phenyl-2,3-Dihydro-1H-Pyrazol-4-Yl)Benzamide
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: FMT, XG7, EDO, DMS |
 |
Pseudomonas Aeruginosa Fabf C164A In Complex With N-(1,5-Dimethyl-3-Oxo-2-Phenyl-2,3-Dihydro-1H-Pyrazol-4-Yl)-2-(4-Methoxyphenoxy)Acetamide
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-03-06 Classification: TRANSFERASE Ligands: XJR, DMS, FMT |
 |
Pseudomonas Aeruginosa Fabf C164A Mutant In Complex With N-(1,5-Dimethyl-3-Oxo-2-Phenyl-2,3-Dihydro-1H-Pyrazol-4-Yl)-3-Methylbutanamide
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-02-21 Classification: TRANSFERASE Ligands: DMS, FMT, ZHX |
 |
Crystal Structure Of F2F-2020209-00X Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2022-11-23 Classification: VIRAL PROTEIN Ligands: 90I, CL, NA |
 |
Crystal Structure Of F2F-2020216-01X Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-11-23 Classification: VIRAL PROTEIN Ligands: 90X, SO4, EDO, CL, NO3, NA |
 |
Crystal Structure Of X77 Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup P2(1)2(1)2(1).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-09-07 Classification: VIRAL PROTEIN Ligands: X77, EDO, CL, DMS, NA |
 |
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup C2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-04-07 Classification: VIRAL PROTEIN Ligands: ALD, EDO, IPA, CL, NA |
 |
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup P1.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-07 Classification: VIRAL PROTEIN Ligands: CL, ALD |

