Search Count: 14
 |
Fucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CA, EDO, FUC |
 |
Allose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, VDS, VDV |
 |
Mannose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, MAN, BMA |
 |
N-Acetyl-Glucosamine-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: NDG, NAG, CA, EDO |
 |
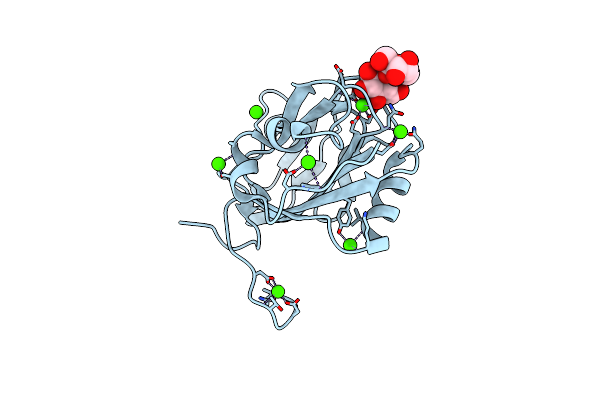
Inositol-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, INS |
 |
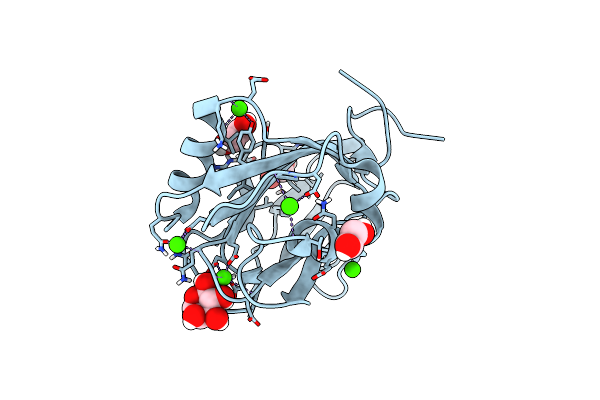
Sucrose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
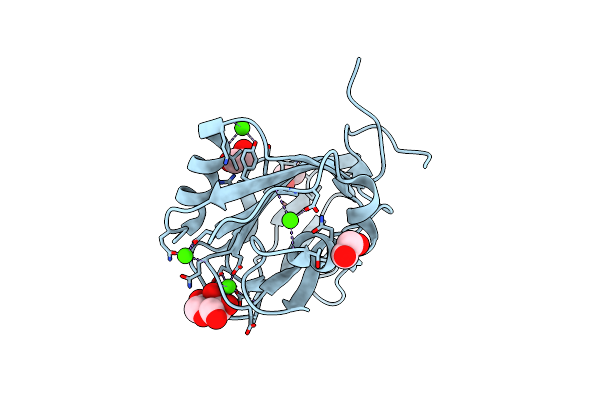
Arabinose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, ARA, EDO |
 |
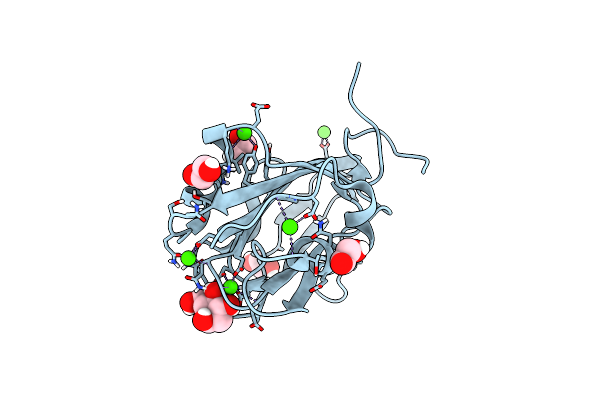
Ribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: RIP, CA, EDO |
 |
2-Deoxy-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: Z61, CA, EDO |
 |
3-O-Methyl-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, 3MG |
 |
2-Deoxyribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: UZJ, CA, EDO |
 |
Trehalose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Galactose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA |
 |
Alpha-Methyl-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, GYP |

