Search Count: 19
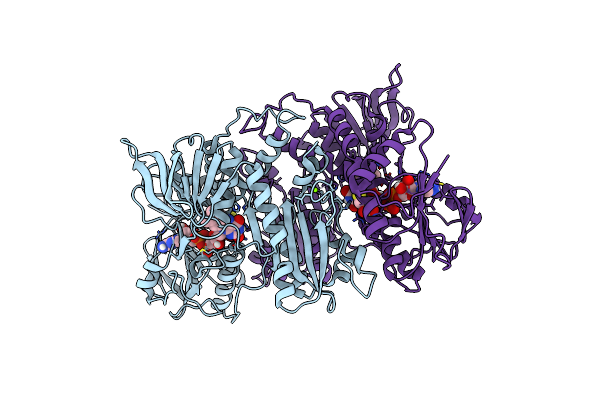 |
Crystal Structure Of F501H/H506E Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: COM, FDA, MG |
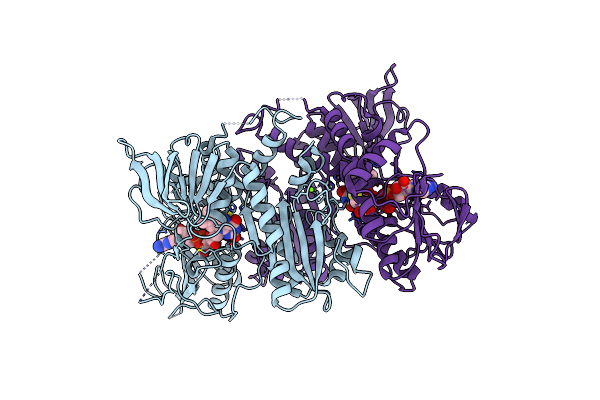 |
Crystal Structure Of F501H Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FDA, COM, MG |
 |
Crystal Structure Of The D100R Multidrug Binding Transcriptional Regulator Lmrr In Complex With Rhodium Bis-Diphosphine Complex
Organism: Lactococcus lactis subsp. cremoris (strain mg1363)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-01 Classification: METAL BINDING PROTEIN Ligands: JY1 |
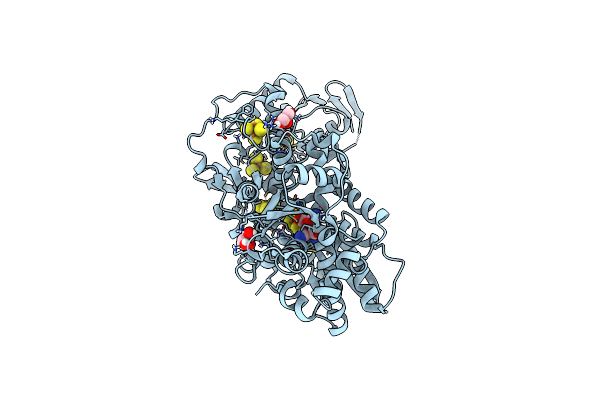 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
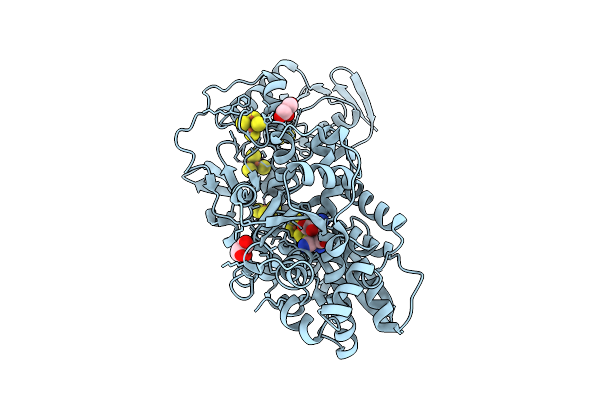 |
Crystal Structure Of [Fefe]-Hydrogenase In The Presence Of 7 Mm Sodium Dithionite
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
 |
Crystal Structure Of [Fefe]-Hydrogenase I (Cpi) Solved With Single Pulse Free Electron Laser Data
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, GOL |
 |
Crystal Structure Of The Multidrug Binding Transcriptional Regulator Lmrr In Complex With Rhodium Bis-Diphosphine Complex
Organism: Lactococcus lactis subsp. cremoris (strain mg1363)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-02-06 Classification: METAL BINDING PROTEIN Ligands: JY1 |
 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-09 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, CLF, FE |
 |
Crystal Structure Of The A-96Gln Mofe Protein Variant In The Presence Of The Substrate Acetylene
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE, ELECTRON TRANSPORT Ligands: C2H, IMD, HCA, ICS, 1CL, FE, MG |
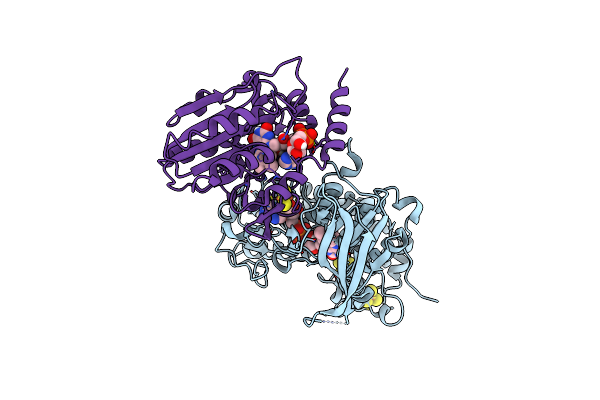 |
Organism: Pyrococcus furiosus com1
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, MG |
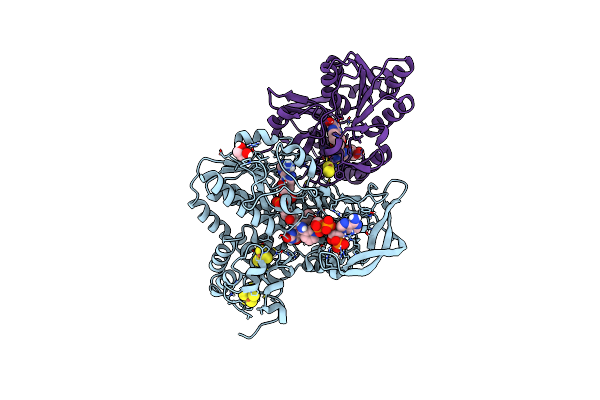 |
Nadp(H) Bound Nadh-Dependent Ferredoxin:Nadp Oxidoreductase (Nfni) From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, NDP, GOL, FES, MG |
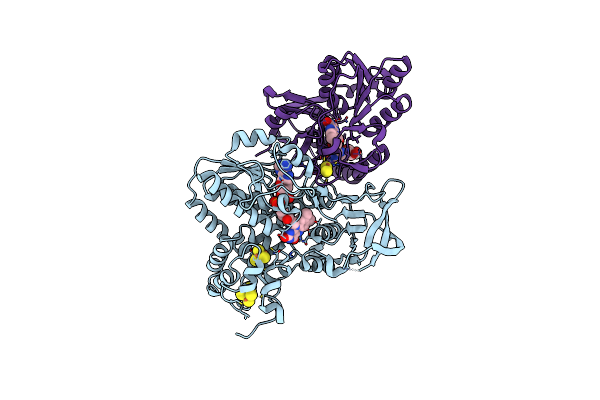 |
Nadh-Dependent Ferredoxin:Nadp Oxidoreductase (Nfni) From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, MG |
 |
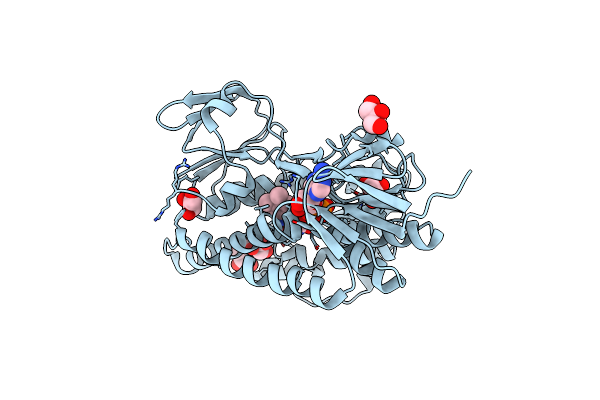
Fe Protein Independent Substrate Reduction By Nitrogenase Variants Altered In Intramolecular Electron Transfer
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-10-14 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, GOL, 1CL, CA, TRS |
 |
Organism: Metallosphaera sedula (strain atcc 51363 / dsm 5348)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2015-09-16 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Chloroflexus aurantiacus (strain atcc 29366 / dsm 635 / j-10-fl)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-07-01 Classification: LYASE Ligands: MG |
 |
Organism: Chloroflexus aurantiacus (strain atcc 29366 / dsm 635 / j-10-fl)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-07-01 Classification: LYASE Ligands: MG, PEP |
 |
Thermostable Enolase From Chloroflexus Aurantiacus With Substrate 2-Phosphoglycerate
Organism: Chloroflexus aurantiacus (strain atcc 29366 / dsm 635 / j-10-fl)
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2015-07-01 Classification: LYASE Ligands: MG, 2PG |
 |
[Fefe]-Hydrogenase Oxygen Inactivation Is Initiated By The Modification And Degradation Of The H Cluster 2Fe Subcluster
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: SF4, CL, ARS |
 |
Recombinant Sperm Whale P6 Myoglobin Solved With Single Pulse Free Electron Laser Data
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2014-11-05 Classification: OXYGEN TRANSPORT Ligands: HEM, SO4 |

