Search Count: 18
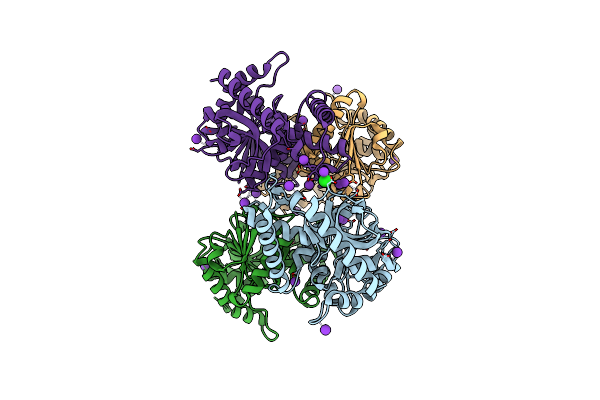 |
Crystal Structure Of Malate Dehydrogenase From Haloarcula Marismortui With Potassium And Chloride Ions
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: OXIDOREDUCTASE Ligands: CL, K |
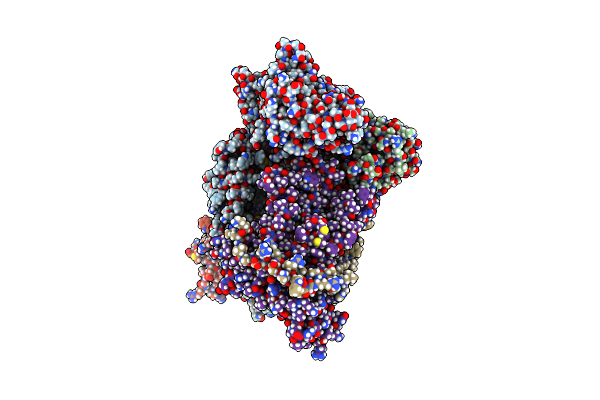 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2016-12-28 Classification: CHAPERONE |
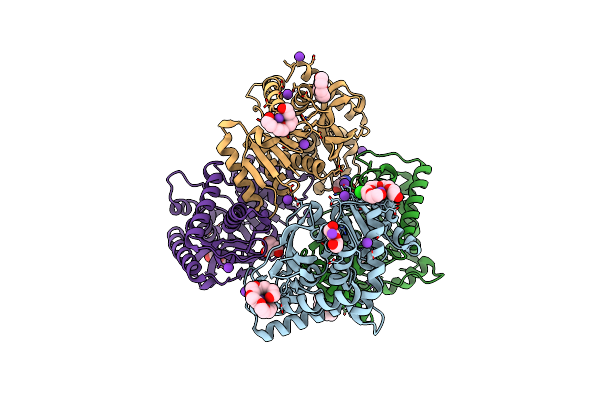 |
1.50 A Resolution Structure Of The Malate Dehydrogenase From Haloferax Volcanii
Organism: Haloferax volcanii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2014-04-16 Classification: OXIDOREDUCTASE Ligands: K, CL, TRS, PGE, 1PE, P33, PG4 |
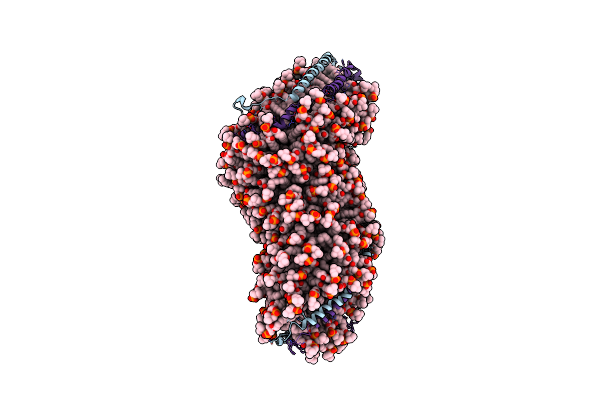 |
Organism: Homo sapiens
Method: SOLUTION SCATTERING Release Date: 2010-04-07 Classification: LIPID BINDING PROTEIN Ligands: POV, CLR |
 |
R207S, R292S Mutant Of Malate Dehydrogenase From The Halophilic Archeon Haloarcula Marismortui (Holoform)
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2009-12-29 Classification: OXIDOREDUCTASE Ligands: NAD, CL, NA |
 |
2.9 A Resolution Structure Of Malate Dehydrogenase From Archaeoglobus Fulgidus In Complex With Nadh
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2009-12-22 Classification: OXIDOREDUCTASE Ligands: NAI, SO4, NA |
 |
2.8 A Resolution Structure Of Malate Dehydrogenase From Archaeoglobus Fulgidus In Complex With Etheno-Nad
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2009-12-22 Classification: OXIDOREDUCTASE Ligands: ENA, SO4 |
 |
Structure Of The Unphotolysed Complex Of Tcache With 1-(2- Nitrophenyl)-2,2,2-Trifluoroethyl-Arsenocholine At 100K
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-20 Classification: HYDROLASE Ligands: CFQ, NAG |
 |
Structure Of The Unphotolysed Complex Of Tcache With 1-(2- Nitrophenyl)-2,2,2-Trifluoroethyl-Arsenocholine After A 9 Seconds Annealing To Room Temperature
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-20 Classification: HYDROLASE Ligands: CL, CFQ, NAG |
 |
Structure Of The Complex Of Tcache With 1-(2-Nitrophenyl)-2,2,2- Trifluoroethyl-Arsenocholine After A 9 Seconds Annealing To Room Temperature, During The First 5 Seconds Of Which Laser Irradiation At 266Nm Took Place
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-11-20 Classification: HYDROLASE Ligands: CL, CFQ, NAG |
 |
Structure Of Native Tcache After A 9 Seconds Annealing To Room Temperature During The First 5 Seconds Of Which Laser Irradiation At 266Nm Took Place
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-20 Classification: HYDROLASE Ligands: NAG |
 |
Torpedo Californica Acetylcholinesterase In Complex With 500Mm Acetylthiocholine
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2006-06-14 Classification: HYDROLASE Ligands: NAG, PGE, AT3, ACE, CL, ACT |
 |
Torpedo Californica Acetylcholinesterase In Complex With 20Mm Acetylthiocholine
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-06-14 Classification: HYDROLASE Ligands: ETM, AT3, NAG, PGE, CL |
 |
Torpedo Californica Acetylcholinesterase In Complex With A Non Hydrolysable Substrate Analogue, 4-Oxo-N,N,N-Trimethylpentanaminium
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-06-14 Classification: HYDROLASE Ligands: NAG, PGE, NWA, CHH |
 |
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-06-14 Classification: HYDROLASE Ligands: ETM, CL, NAG, PGE |
 |
1.95 A Resolution Structure Of (R207S,R292S) Mutant Of Malate Dehydrogenase From The Halophilic Archaeon Haloarcula Marismortui (Holo Form)
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2003-02-06 Classification: OXIDOREDUCTASE Ligands: CL, NAD |
 |
Crystal Structure Of The Wild Type Halophilic Malate Dehydrogenase In The Apo Form
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2000-03-20 Classification: OXIDOREDUCTASE Ligands: CL, NA |
 |
Crystal Structure Of The E267R Mutant Of A Halophilic Malate Dehydrogenase In The Apo Form
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2000-02-04 Classification: OXIDOREDUCTASE Ligands: CL, NA |

