Search Count: 13
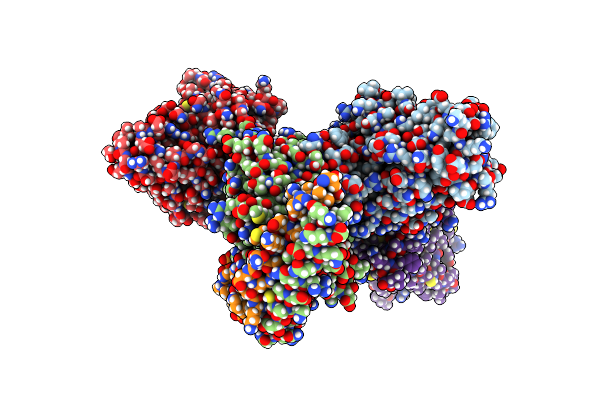 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: SIGNALING PROTEIN Ligands: CLR, ZIT |
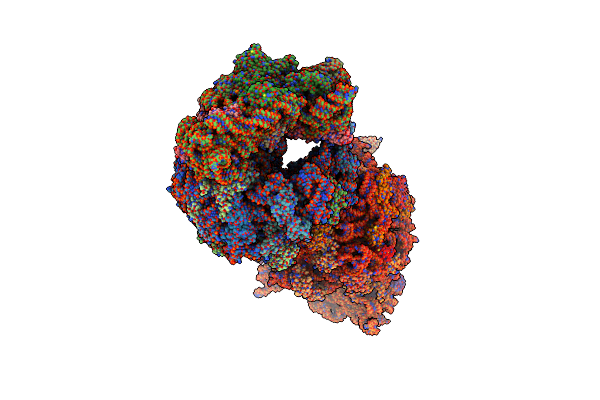 |
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Azithromycin At 2.65A Resolution
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-07-26 Classification: RIBOSOME Ligands: MG, HGR, ZIT, MPD, ARG, ZN, SF4 |
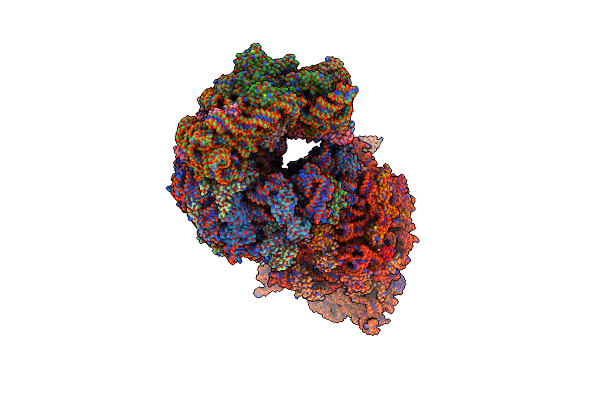 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Azithromycin At 2.50A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: RIBOSOME Ligands: MG, HGR, ZIT, MPD, ARG, ZN, SF4 |
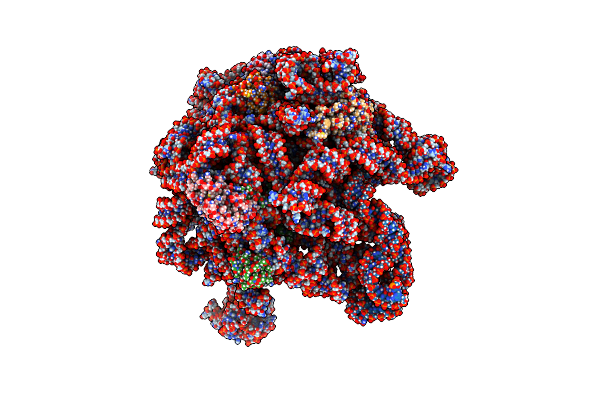 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: RIBOSOME Ligands: MUL, ZIT |
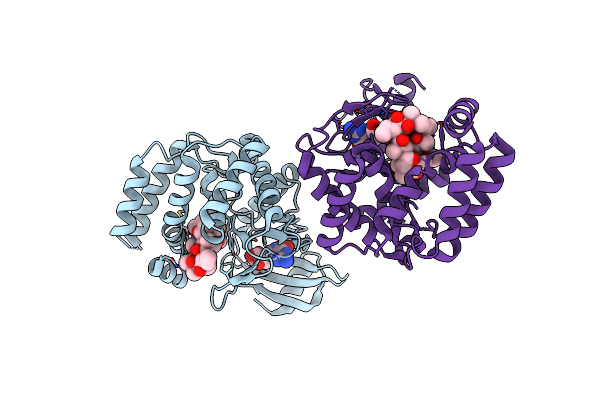 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: GMP, ZIT |
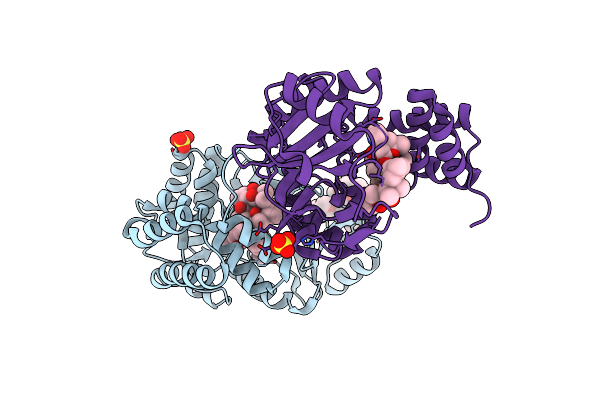 |
Crystal Structure Of Macrolide 2'-Phosphotransferase Mphh From Brachybacterium Faecium In Complex With Azithromycin
Organism: Brachybacterium faecium (strain atcc 43885 / dsm 4810 / ncib 9860)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-08-23 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ZIT, SO4, CL |
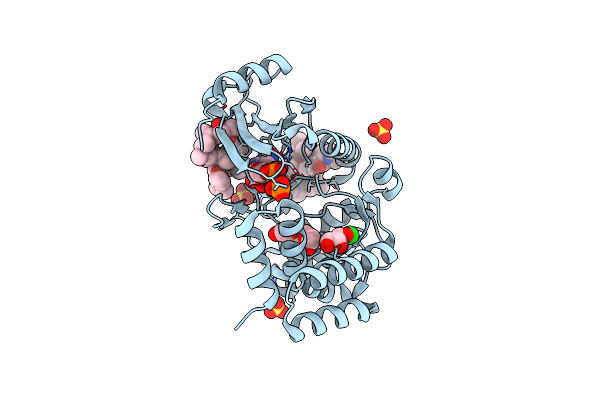 |
Crystal Structure Of Macrolide 2'-Phosphotransferase Mphh From Brachybacterium Faecium In Complex With Gdp
Organism: Brachybacterium faecium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2017-08-16 Classification: TRANSFERASE/ANTIBIOTIC Ligands: MG, GDP, SO4, ZIT, PO4, CL, PGE, GOL |
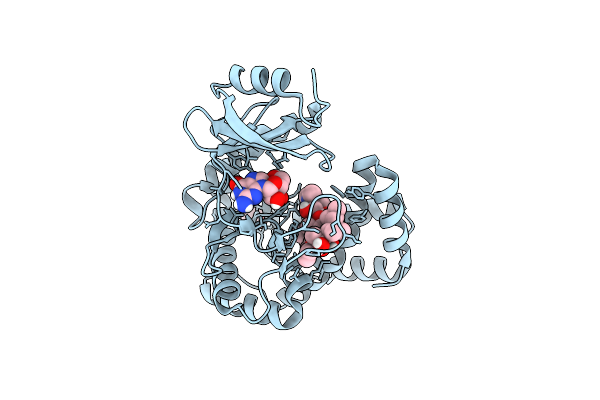 |
Macrolide 2'-Phosphotransferase Type I - Complex With Guanosine And Azithromycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-04-26 Classification: TRANSFERASE/ANTIBIOTIC Ligands: GMP, ZIT, IPA |
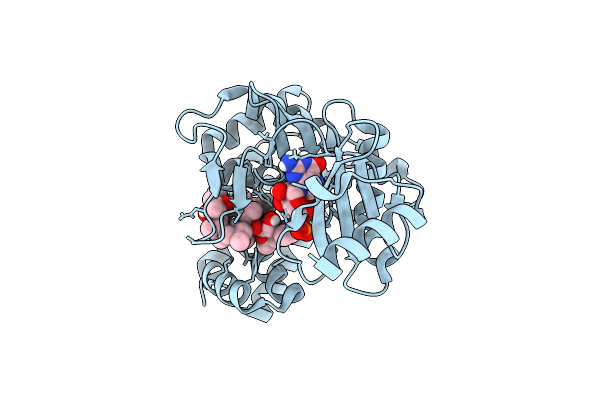 |
Macrolide 2'-Phosphotransferase Type Ii - Complex With Gdp And Azithromycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-04-26 Classification: TRANSFERASE Ligands: MG, CA, GDP, ZIT |
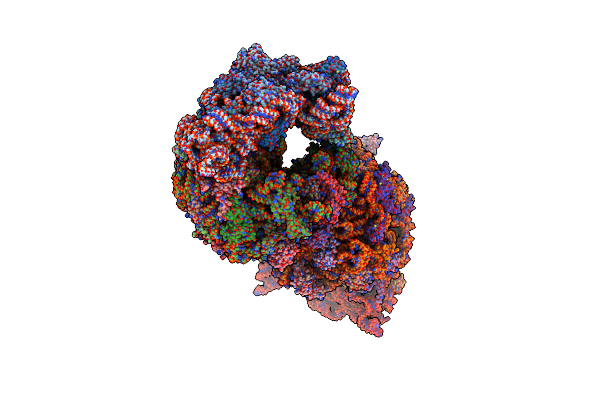 |
Structure Of The Thermus Thermophilus 70S Ribosome Complexed With Azithromycin.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-09 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, ZN, K, ZIT |
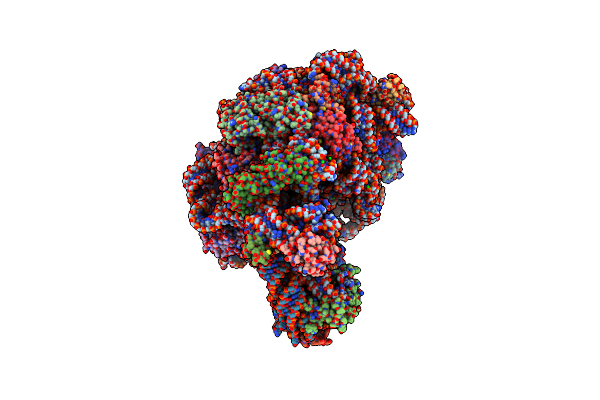 |
Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: ZIT, MG, K, NA, CL, SR, CD |
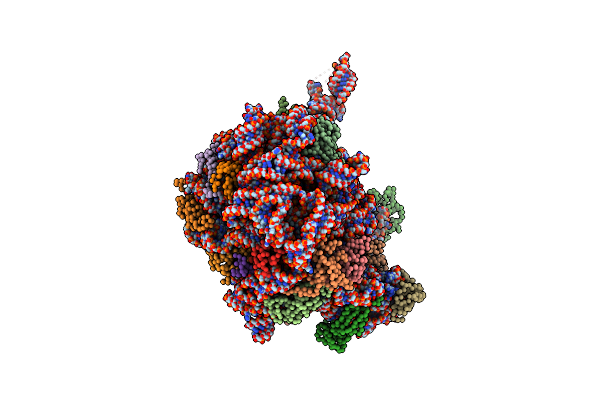 |
Complex Of The Large Ribosomal Subunit From Deinococcus Radiodurans With Azithromycin
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2003-03-18 Classification: RIBOSOME Ligands: ZIT |
 |
Co-Crystal Structure Of Azithromycin Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-07-19 Classification: RIBOSOME Ligands: ZIT, MG, K, NA, CL, CD |

