Search Count: 47
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9 |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9, LMT |
 |
De Novo Designed Cell-Penetrating Peptide Self-Assembly Featuring Distinctive Tertiary Structure
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: DE NOVO PROTEIN |
 |
De Novo Designed Cell-Penetrating Peptide Self-Assembly Featuring Distinctive Tertiary Structure
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-09-11 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
De Novo Designed Cell-Penetrating Peptide Self-Assembly Featuring Distinctive Tertiary Structure
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-09-04 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN Ligands: CXE, KDL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
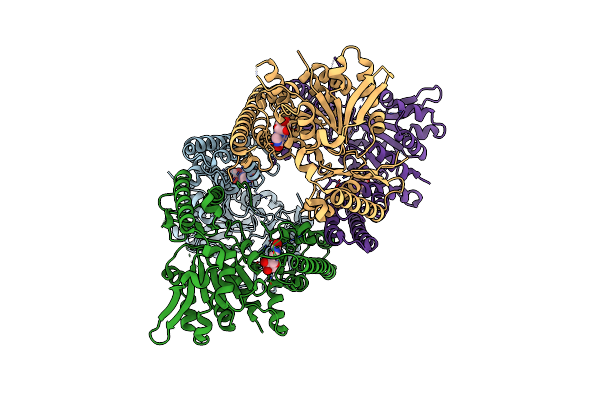 |
Structure And Allosteric Regulation Of The Inosine 5'-Monophosphate-Specific Phosphatase Isn1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NOS |
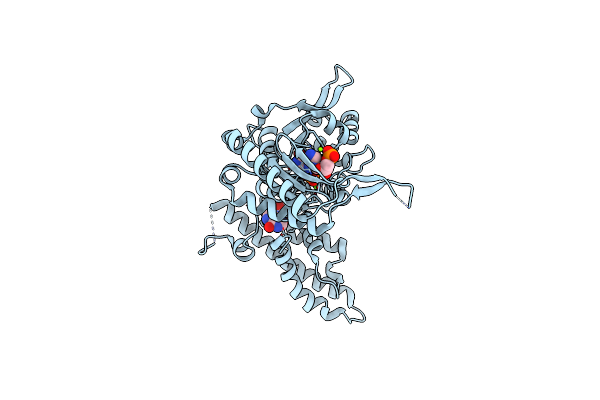 |
Structure And Allosteric Regulation Of The Inosine 5'-Monophosphate-Specific Phosphatase Isn1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NOS, PO4, IMP, MG |
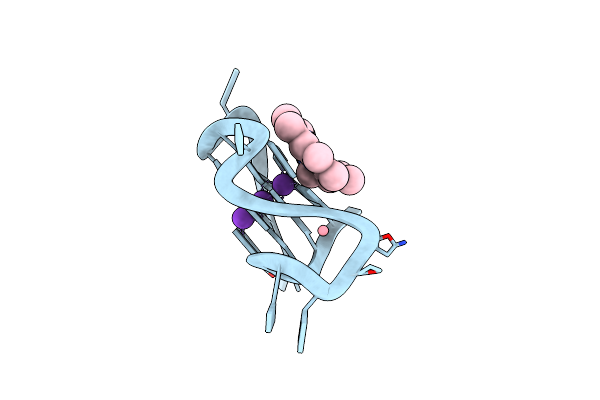 |
Homopurine Parallel G-Quadruplex From Human Chromosome 7 Stabilized By K+ Ions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-12-28 Classification: DNA Ligands: MMP, K, 3CO |
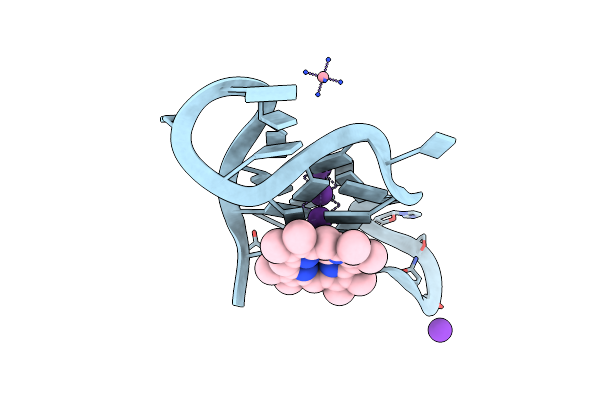 |
Crystal Structure Of A Three-Tetrad, Parallel, And K+ Stabilized Homopurine G-Quadruplex From Human Chromosome 7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-12-28 Classification: DNA Ligands: NCO, K, MMP |
 |
Unliganded Structure Of Pseudouridine Kinase (Puki) From Escherichia Coli Strain B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: K |
 |
Uridine Bound Structure Of Pseudouridine Kinase (Puki) From Escherichia Coli Strain B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: K, URI |
 |
Cytidine Bound Structure Of Pseudouridine Kinase (Puki) From Escherichia Coli Strain B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: CTN |
 |
Pseudouridine Bound Structure Of Pseudouridine Kinase (Puki) S30A Mutant From Escherichia Coli Strain B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: FJF |
 |
Pseudouridine Bound Structure Of Pseudouridine Kinase (Puki) From Escherichia Coli Strain B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: FJF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-02-17 Classification: TRANSFERASE Ligands: 3JW |

