Search Count: 56
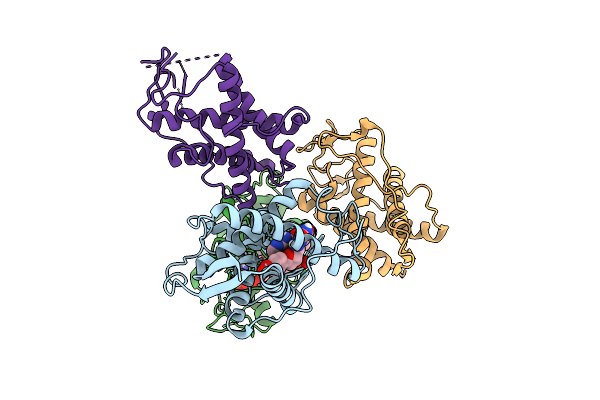 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Dantrolene And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: U1C, ACP |
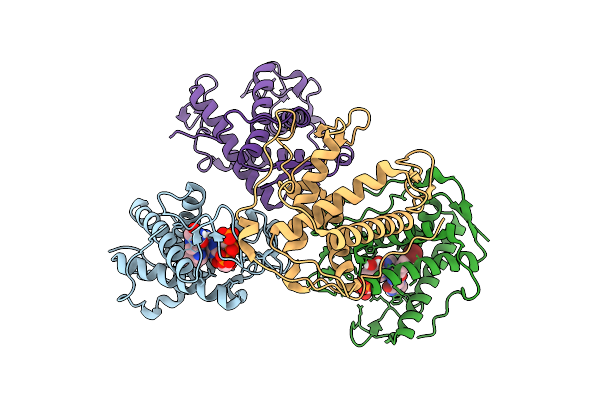 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Azumolene And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ACP, A1L7B |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: GOL |
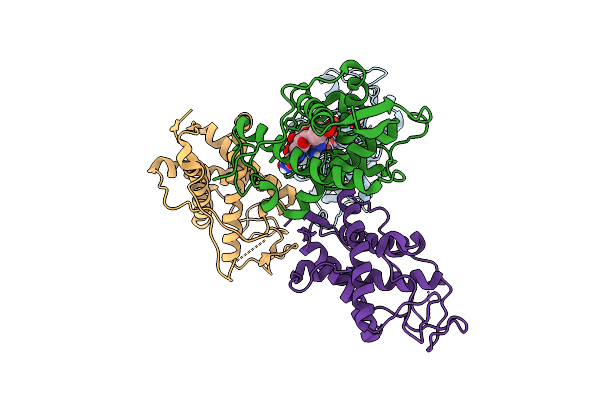 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Dantrolene And Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ADP, U1C |
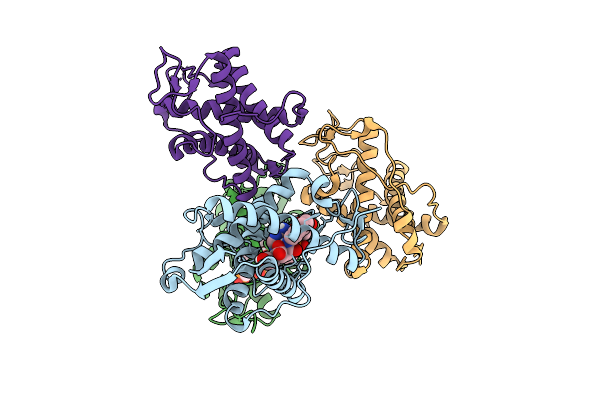 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain Complex With Ri-2 And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ACP, A1L7G |
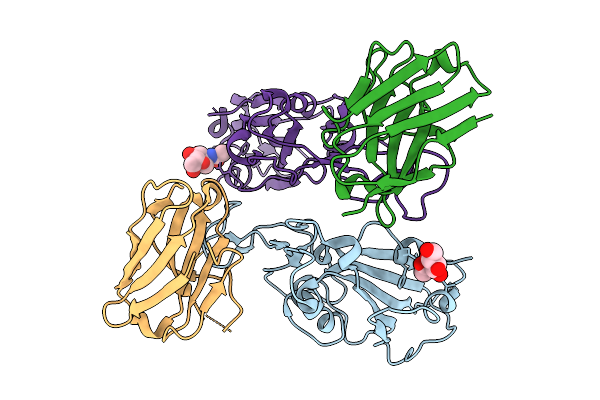 |
The Complex Structure Of Sars-Cov-2 Rbd And Llama Single-Domain Antibody S4
Organism: Severe acute respiratory syndrome coronavirus 2, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: A1LVX, ZN, CA, ATP, CFF |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: ZN, CA, ATP, CFF, A1LV1 |
 |
Organism: Oryctolagus cuniculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: CA, ZN, ATP, CFF |
 |
Organism: Oryctolagus cuniculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: ZN, CA, ATP, CFF |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: F0U, ZN, CA, ATP, CFF |
 |
Crystal Structure Of The Rgs-Homologous Domain Of Axin In Complex With Lz-22Na
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-12-13 Classification: SIGNALING PROTEIN Ligands: I5L |
 |
Organism: Phytophthora sojae strain p6497
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: TRANSFERASE |
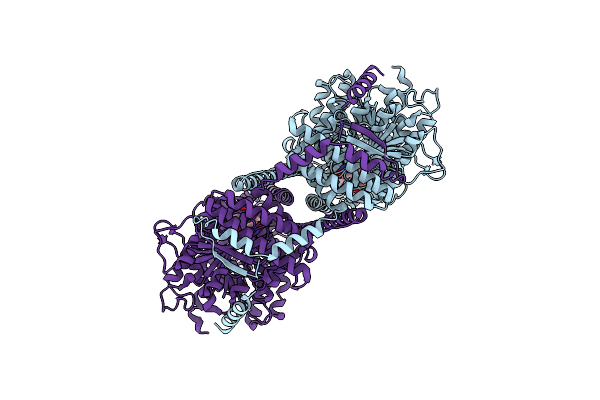 |
Cryoem Structure Of Chitin Synthase 1 Mutant E495A From Phytophthora Sojae Complexed With Udp-Glcnac
Organism: Phytophthora sojae strain p6497
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: UD1, MN |
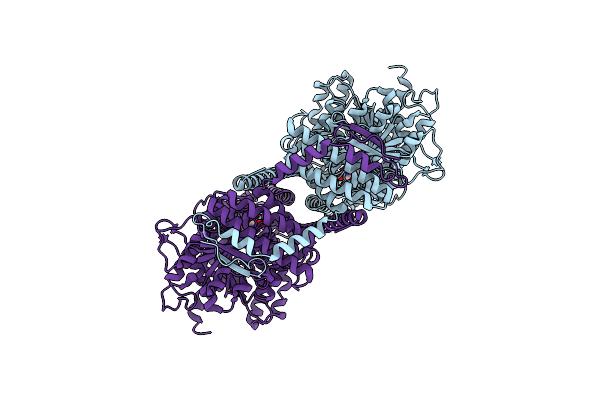 |
Cryoem Structure Of Chitin Synthase 1 From Phytophthora Sojae Complexed With Nikkomycin Z
Organism: Phytophthora sojae strain p6497
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: BGI |
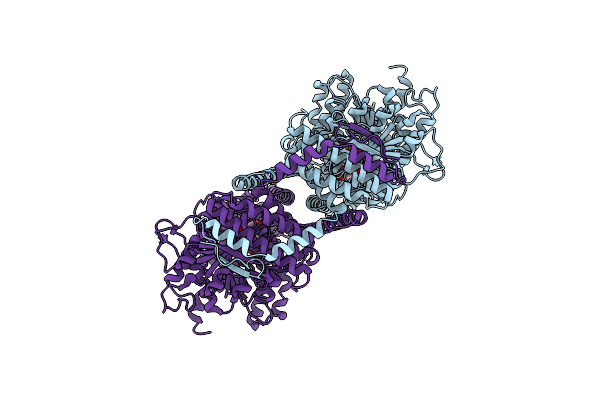 |
Cryoem Structure Of Chitin Synthase 1 From Phytophthora Sojae Complexed With The Nascent Chitooligosaccharide
Organism: Phytophthora sojae strain p6497
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: UDP, MN |
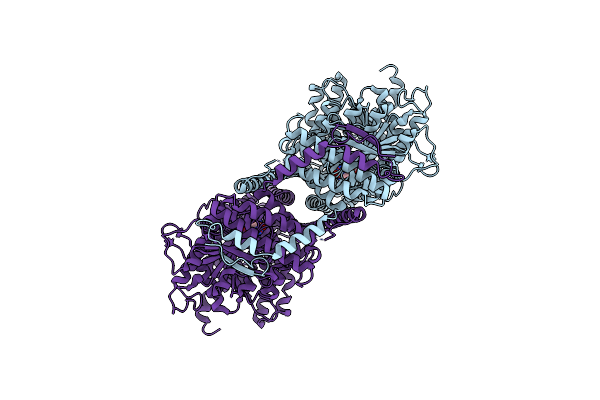 |
Cryoem Structure Of Chitin Synthase 1 From Phytophthora Sojae Complexed With Udp
Organism: Phytophthora sojae strain p6497
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: UDP, MG |
 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: ZN, DPO, GLY |
 |
Anaerobic Hydroxyproline Degradation Involving C-N Cleavage By A Glycyl Radical Enzyme
Organism: Clostridiales bacterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-01 Classification: LYASE Ligands: UY7 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-04-27 Classification: TRANSFERASE Ligands: ANP, MG |

