Search Count: 116
 |
Ros-Sensing Transcription Factor Ychj Regulates The Rssb-Rpos Pathway To Protect Salmonella Against Oxidative Attack By Macrophages
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Vicugna pacos, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: ANTIVIRAL PROTEIN |
 |
Structural Analysis Of Autophagy-Related Protein 8A In Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-03-26 Classification: LIPID TRANSPORT |
 |
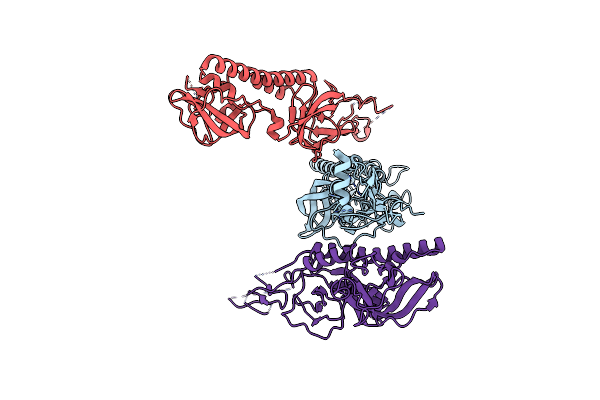
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSCRIPTION/DNA |
 |
Organism: Lyssavirus mokola
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: VIRAL PROTEIN |
 |
Organism: Ikoma lyssavirus
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nynylyrll(Eg.5.1)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nydywyrlf(Ba.2.86)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nynylyrlf(Prototype)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
The Structure Of Hla-A*0201 Complex With Peptide From Sars-Cov-2 N222-230 Llldrlnkl(Ba.2.86/Jn.1)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nydywyrsf(Jn.1)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nynyryrlf(Delta/Ba.5.2)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
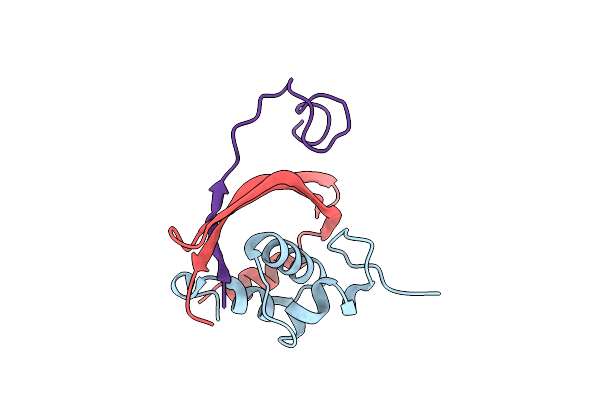
The Structure Of Hla-A*2402 Complex With Peptide From Sars-Cov-2 S448-456 Nynyqyrlf(Ba.2.12.1)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-01-29 Classification: IMMUNE SYSTEM |
 |
A Ros-Sensing Transcription Factor Promotes Rpos Accumulation To Resist Oxidative Stress
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-11-13 Classification: IMMUNE SYSTEM Ligands: 6IL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-09-25 Classification: ISOMERASE Ligands: A1D9K, SO4, PE3 |

