Search Count: 90
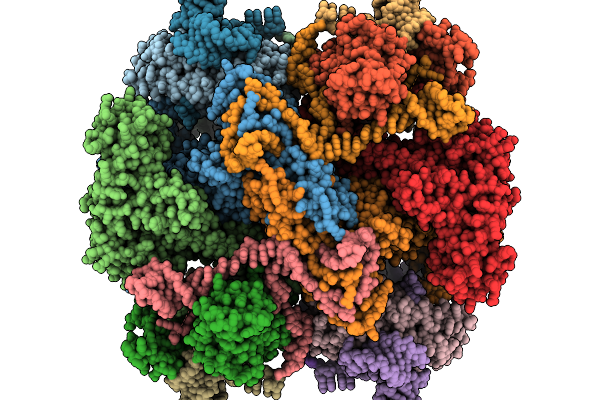 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA |
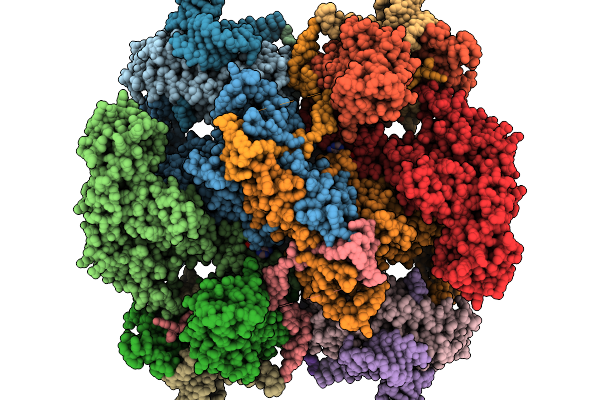 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTITOXIN/DNA/RNA Ligands: ATP |
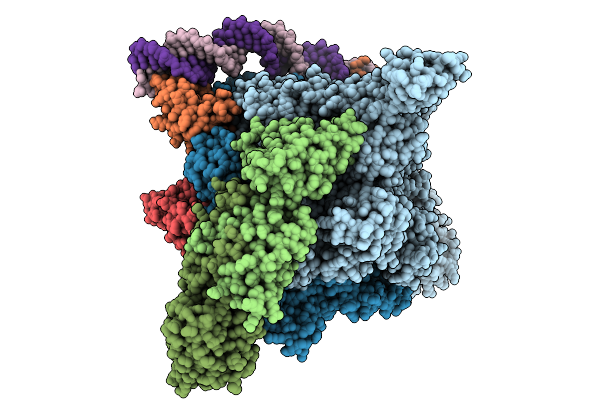 |
Organism: Escherichia coli, Escherichia phage phieco32
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSCRIPTION |
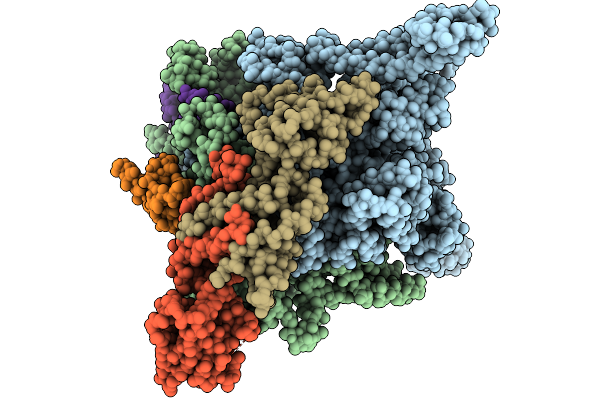 |
Organism: Escherichia coli, Escherichia phage phieco32
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: IMMUNE SYSTEM Ligands: A1BLD |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNE SYSTEM |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNE SYSTEM |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNOSUPPRESSANT |
 |
The Cryo-Em Structure Of Anti-Phage Defense Associated Dsr2 Tetramer Bound With Two Dsad1 Inhibitors (Same Side)
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNOSUPPRESSANT |
 |
The Cryo-Em Structure Of Anti-Phage Defense Associated Dsr2 Tetramer Bound With Two Dsad1 Inhibitors (Opposite Side)
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNOSUPPRESSANT |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: IMMUNE SYSTEM Ligands: NAD |
 |
Cryo-Em Structure Of Staphylococcus Aureus Siga-Dependent Rnap-Promoter Open Complex
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: TRANSCRIPTION/DNA |

