Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Pectobacterium atrosepticum scri1043
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
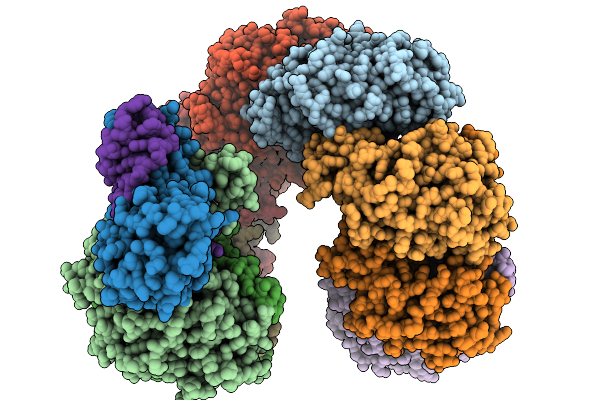
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens dsm 198
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: ITN |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
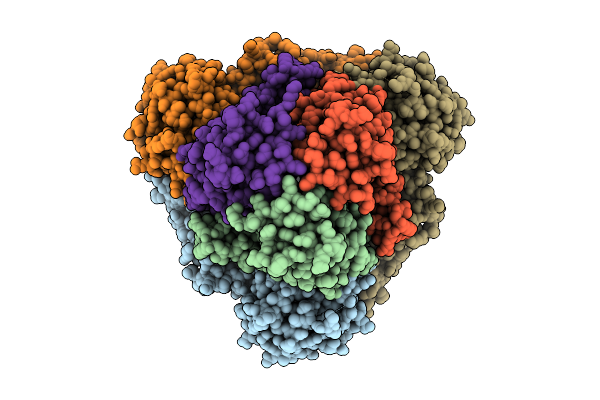
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
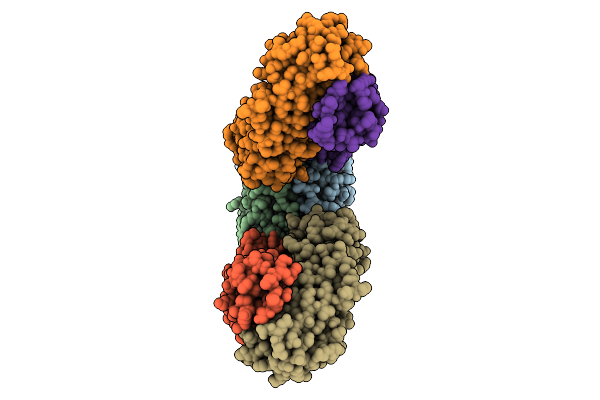
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
 |
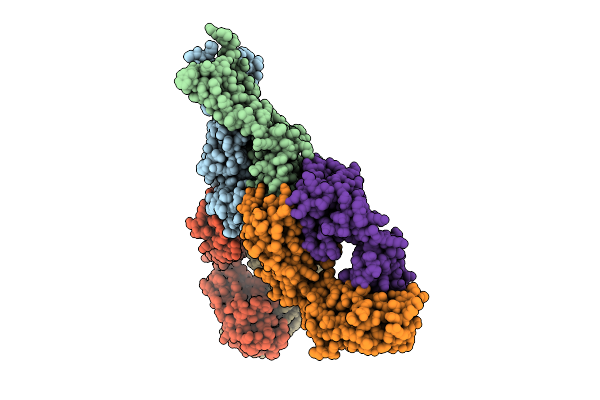
Organism: Sars-cov-2 pseudovirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM/INHIBITOR Ligands: A1AE8 |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM/INHIBITOR |
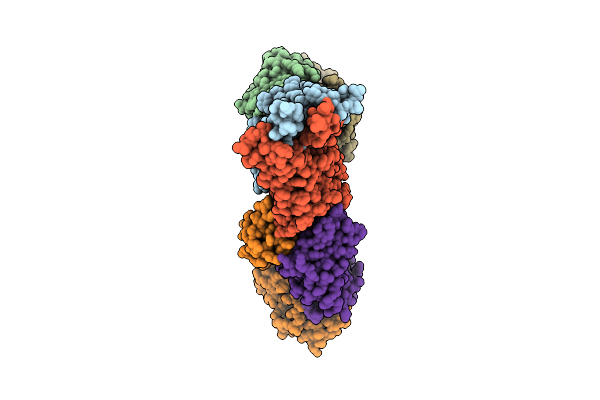 |
Organism: Middle east respiratory syndrome-related coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM/INHIBITOR |
 |
Cryoem Structure Of Respiratory Syncytial Virus Polymerase In Complex With Novel Non-Nucleoside Inhibitor Compound 16
Organism: Human respiratory syncytial virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN,TRANSFERASE/INHIBITOR |
 |
Cryoem Map Of Respiratory Syncytial Virus Polymerase With Non-Nucleoside Inhibitor Compound 21
Organism: Human respiratory syncytial virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/INHIBITOR |
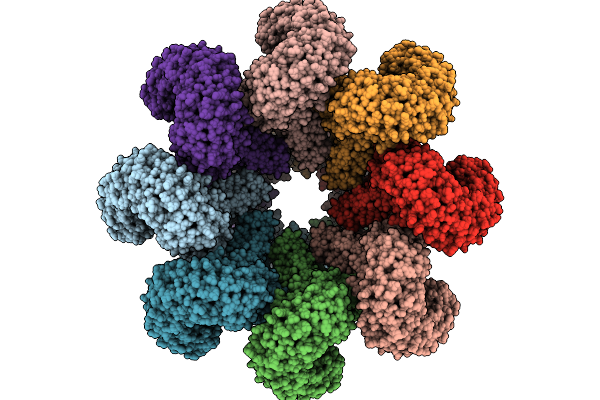 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
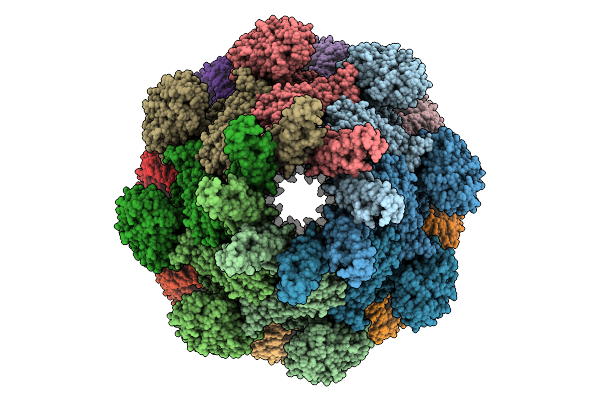 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
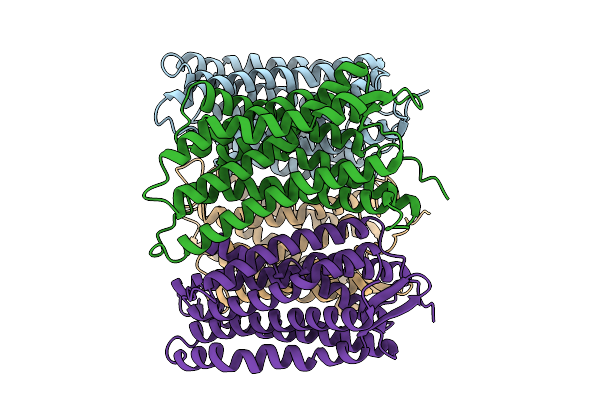 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: A1EF4, OGA, FE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Uq1-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, UQ1, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Pydiflumetofen-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EE4, PEE |