Search Count: 195
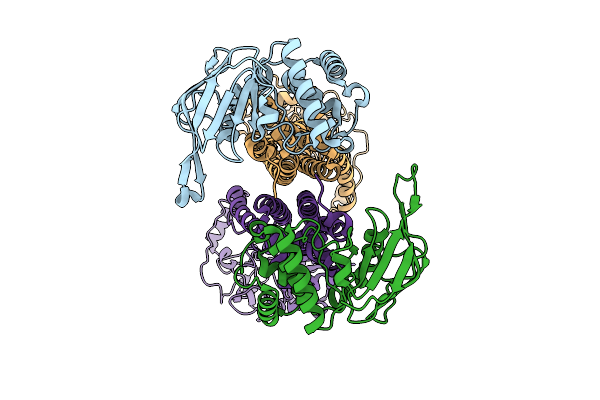 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
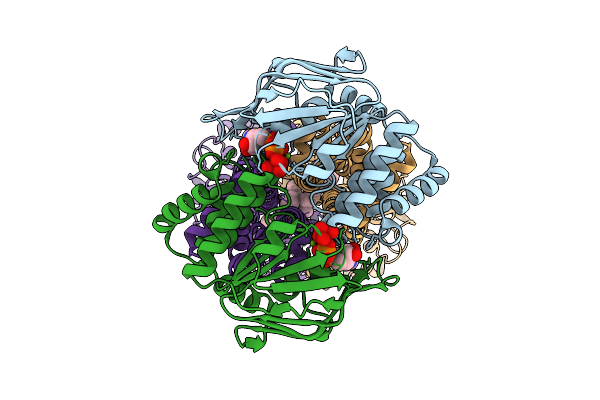 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: ADP, VO4, MG, UNL |
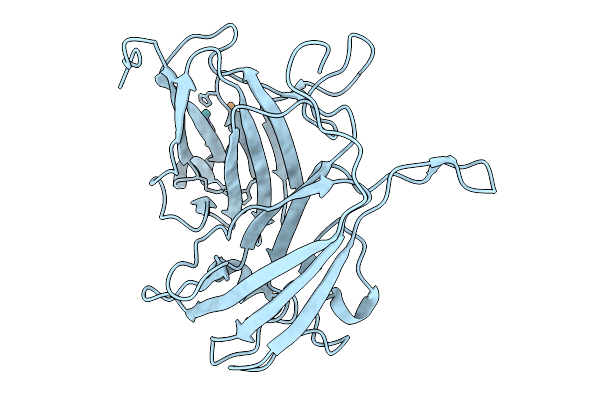 |
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: OXIDOREDUCTASE Ligands: CU, RU |
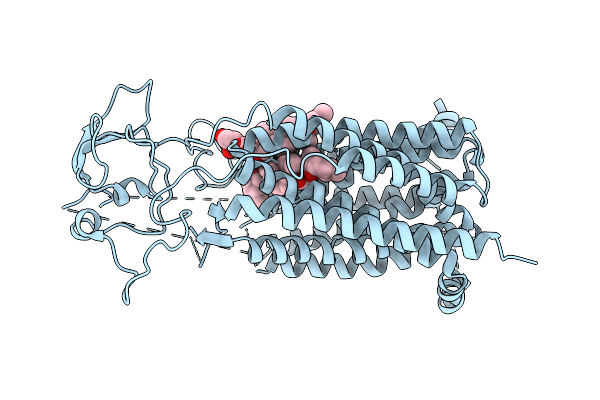 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: AJP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTITUMOR PROTEIN Ligands: A1EG3 |
 |
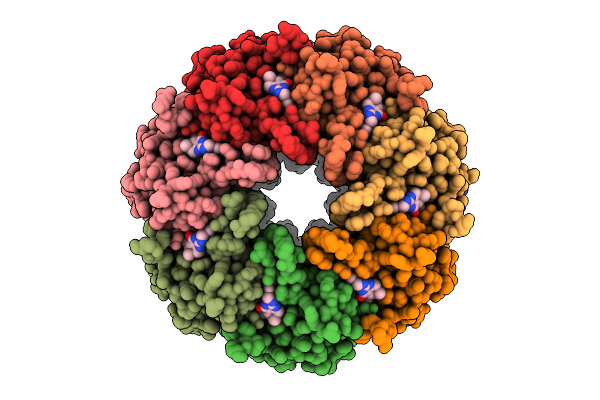
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: A1EBP, GOL, BEZ, PDO, BU1, PGO |
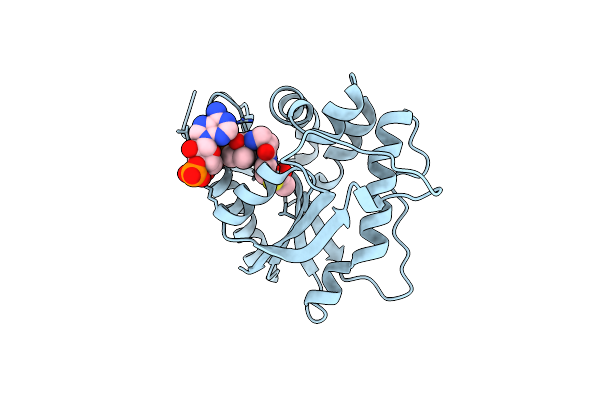 |
Crystal Structure Of Drosophila Melanogaster D46A Dopamine N-Acetyltransferase In Complex With Acetyl-Coa
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: ACO |
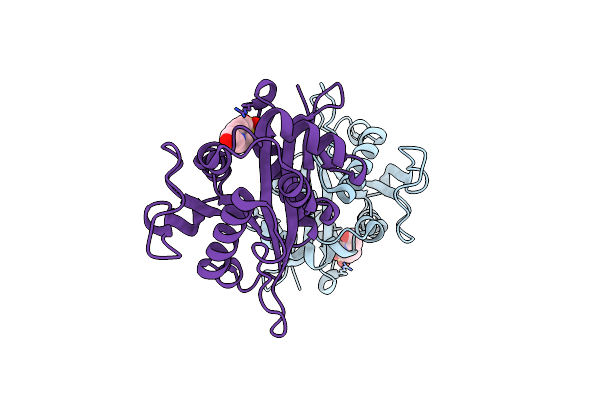 |
Crystal Structure Of Drosophila Melanogaster D46A Dopamine N-Acetyltransferase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: EPE |
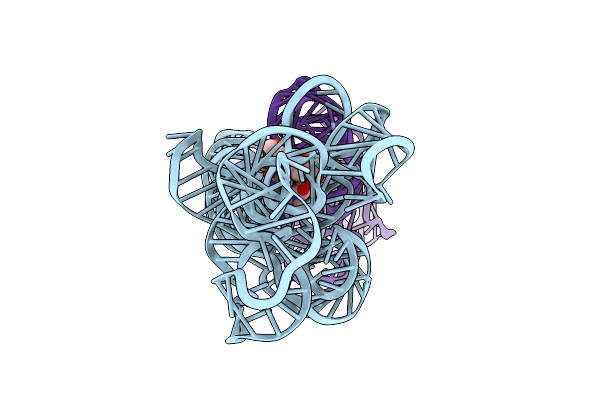 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: RNA Ligands: B1Z |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: RNA Ligands: B1Z |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: RNA Ligands: B1Z |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: RNA Ligands: B1Z |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-06-21 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-06-21 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-06-21 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-06-21 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-05-17 Classification: RNA Ligands: IRI, PGE, PG4 |
 |
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: SO4, MPG |
 |
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-11-09 Classification: OXIDOREDUCTASE Ligands: CO |

