Search Count: 25
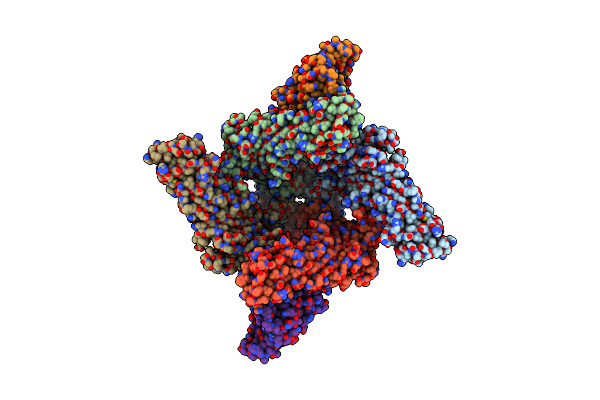 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: P5S, U6L, GDP, MG |
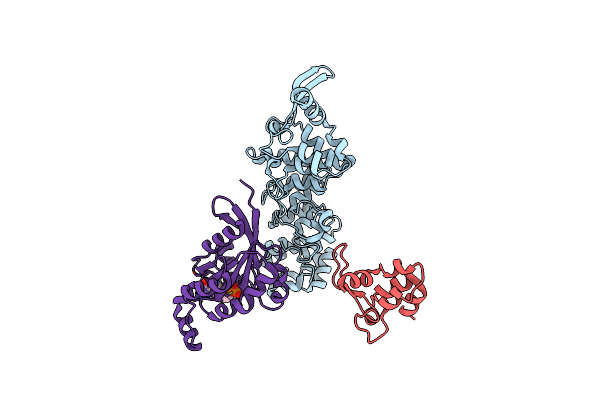 |
Cryo-Em Structure Of Human Trpv4 Intracellular Domain In Complex With Gtpase Rhoa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: GDP, MG |
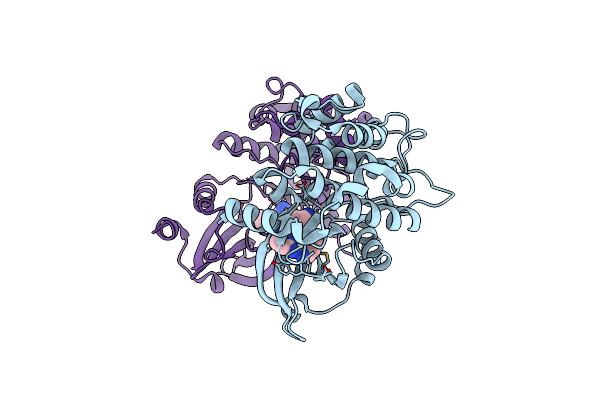 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2P |
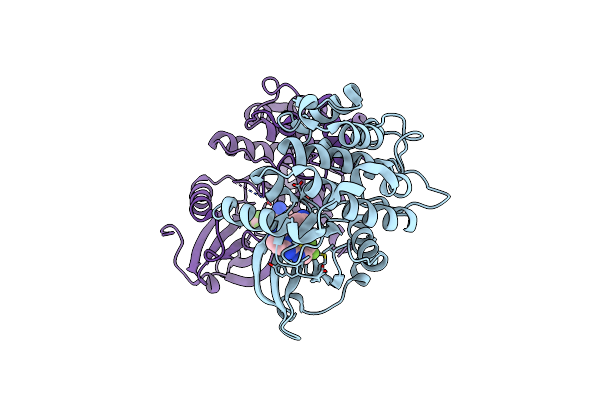 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2R, EDO |
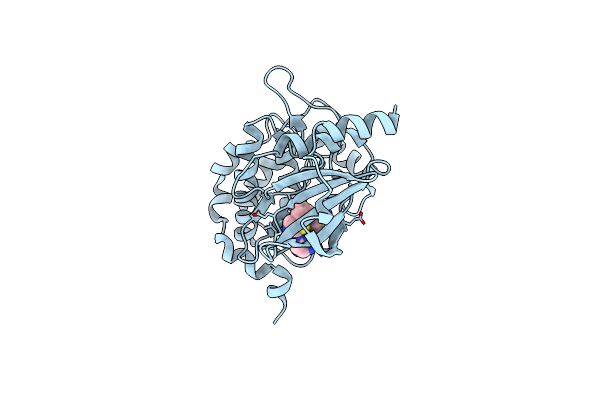 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: A1A2P |
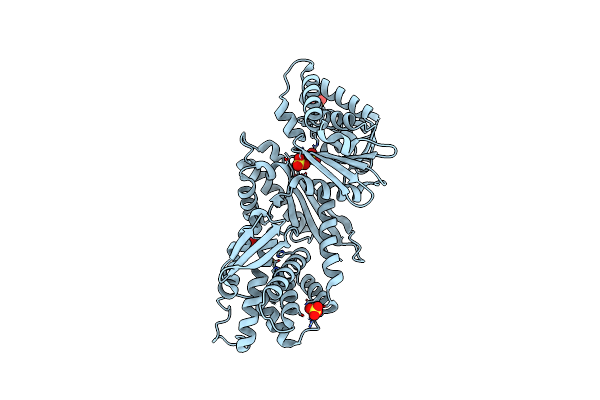 |
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: SO4, MG |
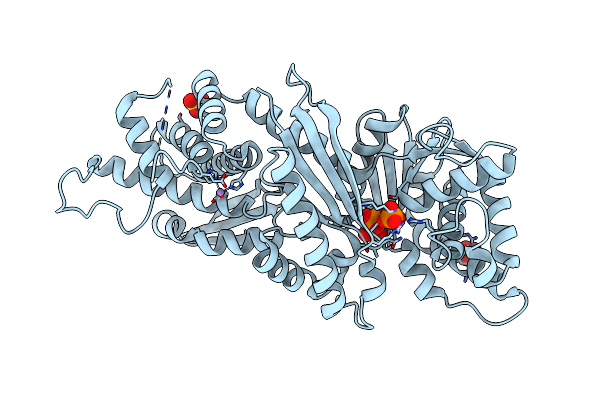 |
Crystal Structure Of Deinococcus Radiodurans Exopolyphosphatase Complexed With Pyrophosphate
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: PPV, MN, SO4, PO4 |
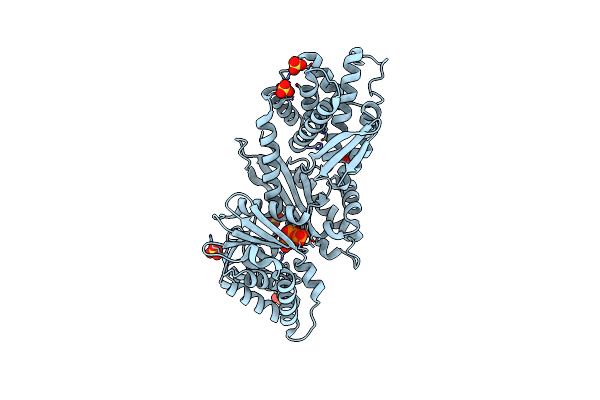 |
Crystal Structure Of Deinococcus Radiodurans Exopolyphosphatase Complexed With P5
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: UG3, SO4, MG |
 |
Crystal Structure Of Deinococcus Radiodurans Exopolyphosphatase In The Presence Of Pi
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: PO4, K |
 |
Crystal Structure Of Deinococcus Radiodurans Exopolyphosphatase E114A Mutant
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
Crystal Structure Of N-Terminal Domain Of Exopolyphosphatase From Deinococcus Radiodurans
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-05-15 Classification: HYDROLASE |
 |
Organism: Klebsiella pneumoniae subsp. pneumoniae 1158
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: GOL, K, MG |
 |
Organism: Acinetobacter baumannii 15827
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-05-15 Classification: HYDROLASE |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1), Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: GENE REGULATION Ligands: MG |
 |
Organism: Thermococcus thioreducens, Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, MG |

