Search Count: 648
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: 8K3 |
 |
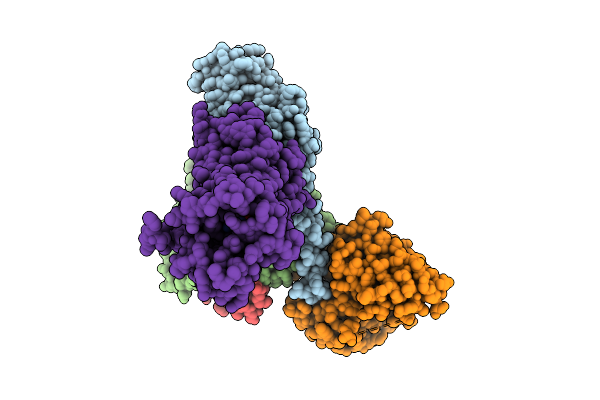
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 1)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
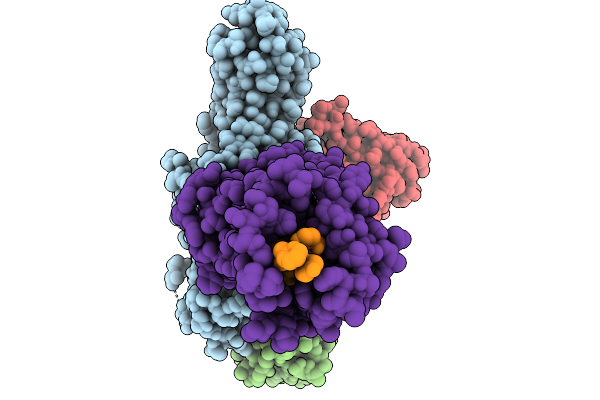
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 2)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
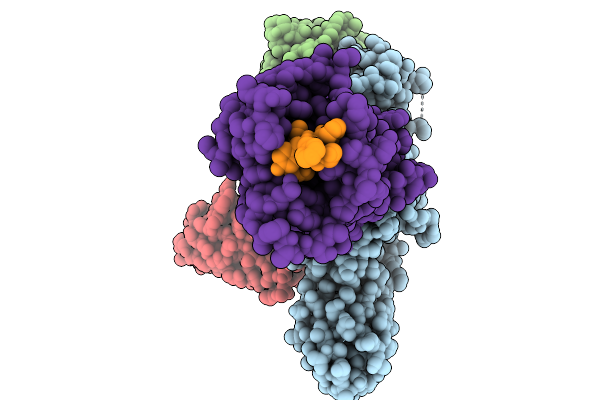
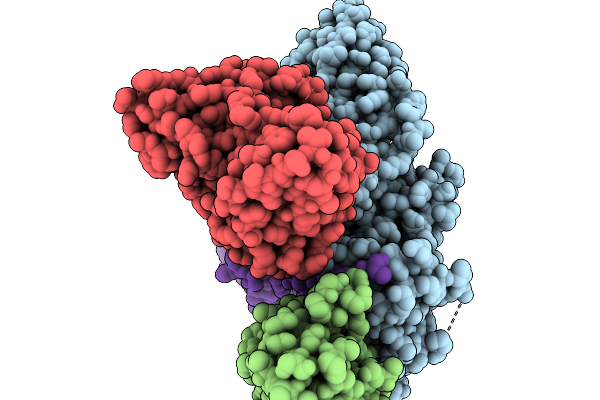
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 3)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
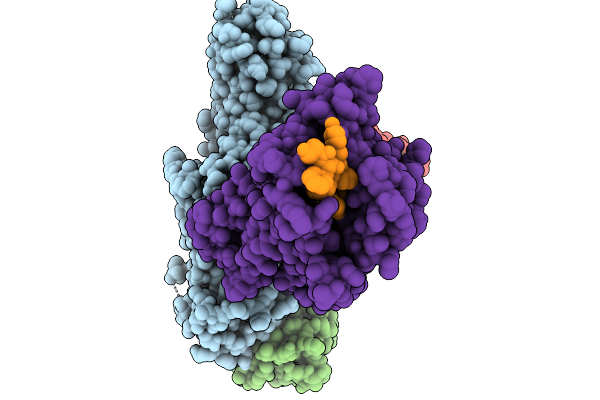
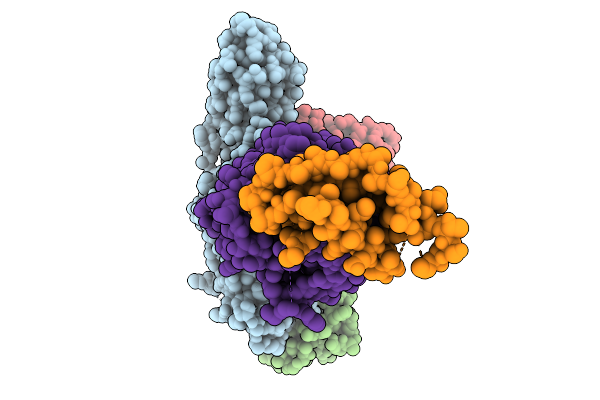
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 4)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
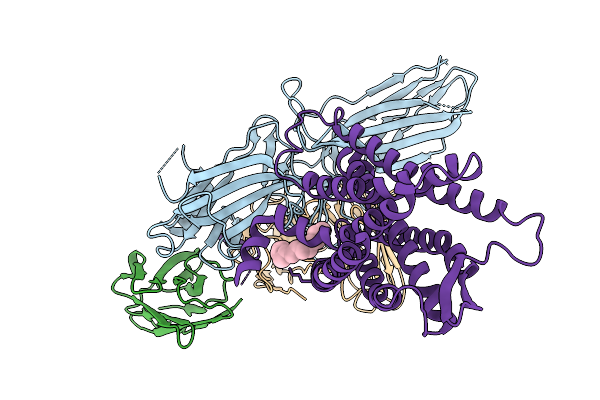
Composite Map Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 2
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
 |
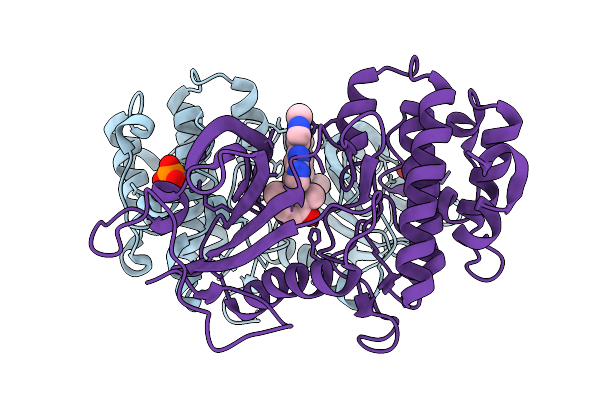
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: PAM |
 |
Discovery Of A Bifunctional Pkmyt1-Targeting Protac Empowered By Ai-Generation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EMX, PO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ELECTRON TRANSPORT Ligands: HEC, ZN, PEG, 1PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9 |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9, LMT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Cryo-Em Structure Of Human Separase Bound To Phosphorylated Scc1 (310-550 Aa)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Cryo-Em Structure Of Human Separase Bound To Phosphorylated Scc1 (100-320 Aa)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiose
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiitol
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: MTL, BMA, CL, PO4 |

