Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Thermothielavioides terrestris (strain atcc 38088 / nrrl 8126)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, MG |
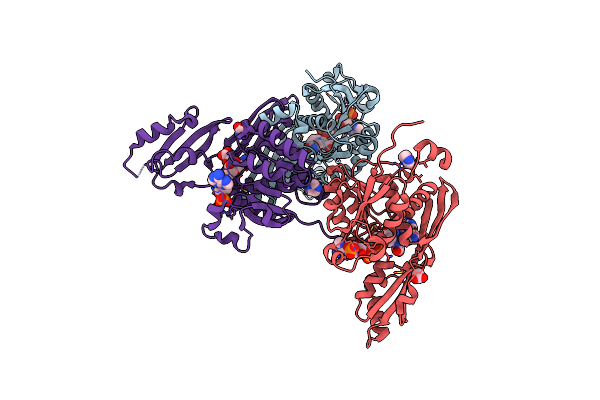 |
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Bacillus Thermozeamaize Mutant M9 Complexed With Nadp+
Organism: Bacillus thermozeamaize
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: NAP, OXM, IMD, ACY |
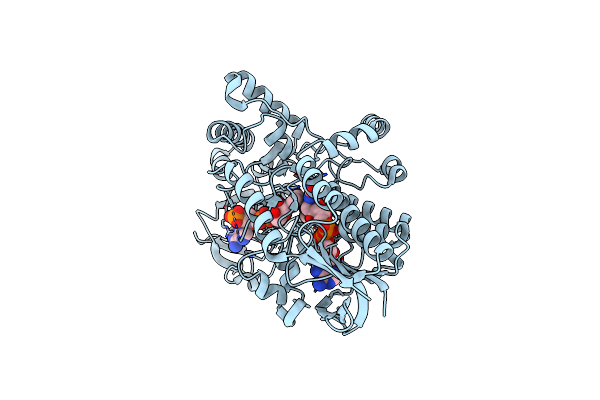 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
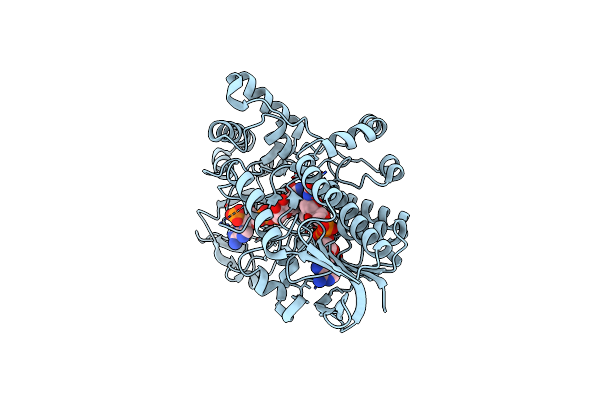 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
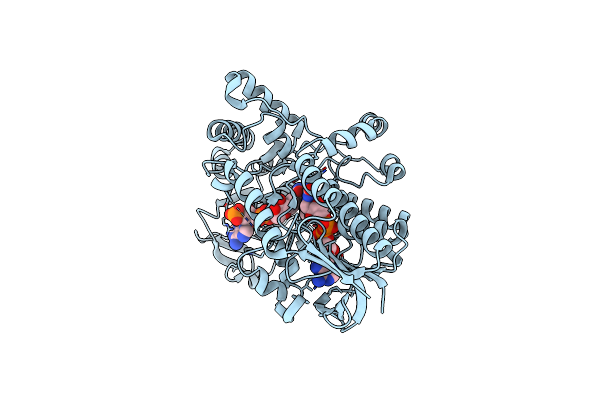 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NDP, FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NAI, FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Crystal Structures Of Meso-Diaminopimelate Dehydrogenase From Bacillus Thermozeamaize
Organism: Bacillus thermozeamaize
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde From The Cyanohydrin Cleavage
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: LYASE Ligands: NAG, FQ3, FAD, PGE, PEG, BCN |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant From Prunus Communis Mutant L331A Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde (Form A)
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: LYASE Ligands: NAG, FQ3, FAD, EDO, PEG, BCN |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde (Form B)
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: LYASE Ligands: NAG, FQ3, PGE, FAD, PEG, EDO |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With 4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: LYASE Ligands: NAG, FQ6, FAD, PEG, PO4 |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With 2-Methyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: LYASE Ligands: FQ9, FAD, NAG, PEG, DMS |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A/S333V/P340L From Prunus Communis
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: LYASE Ligands: FAD, PEG, NAG, EDO |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A/S333V/P340L From Prunus Communis Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde (Catalytic Conformation)
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: LYASE Ligands: FQ3, FAD, NAG, PO4 |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A/S333V/P340L From Prunus Communis Complexed With 2,2-Dimethyl-4H-Benzo[D][1,3]Dioxine-6-Carbaldehyde (Noncatalytic Conformation)
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: LYASE Ligands: FQ3, FAD, NAG, PEG, PO4 |
 |
Endo-Deglycosylated Hydroxynitrile Lyase Isozyme 5 Mutant L331A From Prunus Communis Complexed With (R)-2-(2,2-Dimethyl-4H-Benzo[D][1,3]Dioxin-6-Yl)-2-Hydroxyacetonitrile
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Release Date: 2024-06-05 Classification: LYASE Ligands: E2Y, NAG, FAD, PEG, EDO |
 |
Formate Dehydrogenase Wild-Type Enzyme From Candida Dubliniensis Complexed With Nadh
Organism: Candida dubliniensis (strain cd36 / atcc mya-646 / cbs 7987 / ncpf 3949 / nrrl y-17841)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: NAI, MG |