Search Count: 680
 |
Organism: Thermothielavioides terrestris (strain atcc 38088 / nrrl 8126)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, MG |
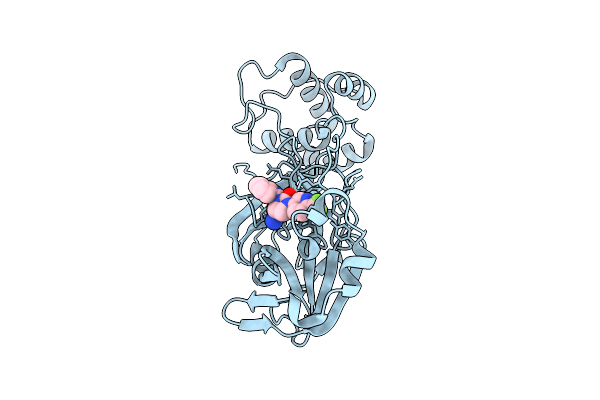 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
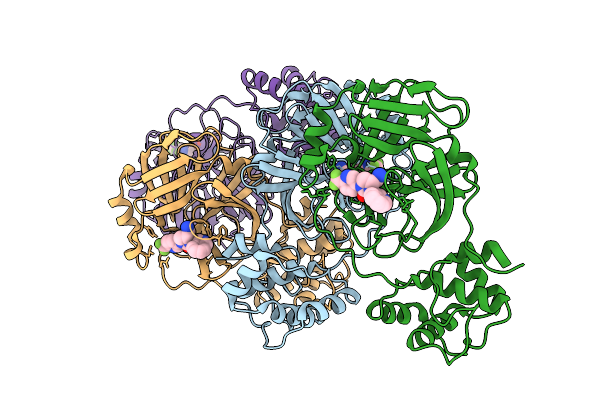 |
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
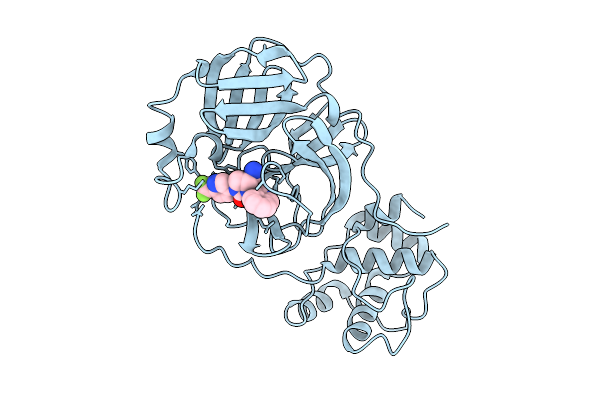 |
Organism: Human coronavirus oc43
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: SF4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
Organism: Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
 |
Organism: Human herpesvirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Mus musculus, Cercopithecine herpesvirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Suid alphaherpesvirus 1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Human herpesvirus 3, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Rocahepevirus ratti, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Hepatitis E Virus Capsid Protein E2S Domain (Genotype I) In Complex With Fab C6
Organism: Hepatitis e virus genotype 1 (isolate human/burma), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Hepatitis E Virus Capsid Protein E2S Domain (Genotype Iv) In Complex With Fab C6
Organism: Hepatitis e virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

