Search Count: 254
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT |
 |
Organism: Mus musculus, Synthetic constrcut
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-11-05 Classification: TRANSFERASE |
 |
Integrin Aiibb3 Bent Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
 |
Integrin Aiibb3 Intermediate Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
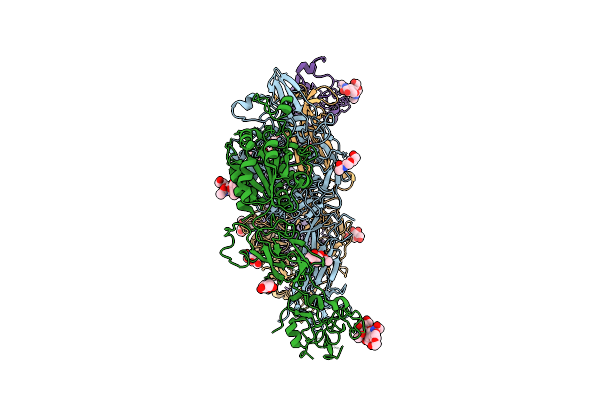 |
Integrin Aiibb3 Dimer Conformation From Human Platelet Membrane Crude Preparation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: BLOOD CLOTTING Ligands: CA, NAG, MG |
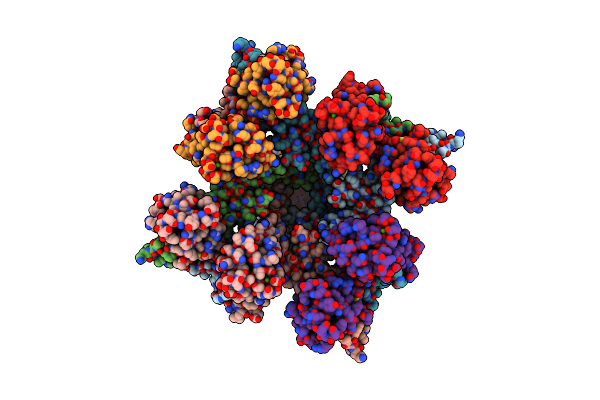 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
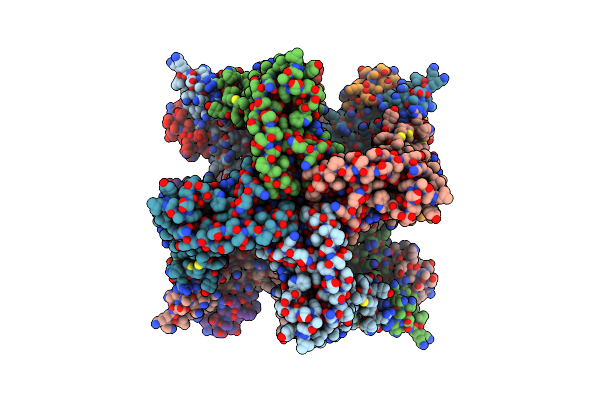 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, CA |
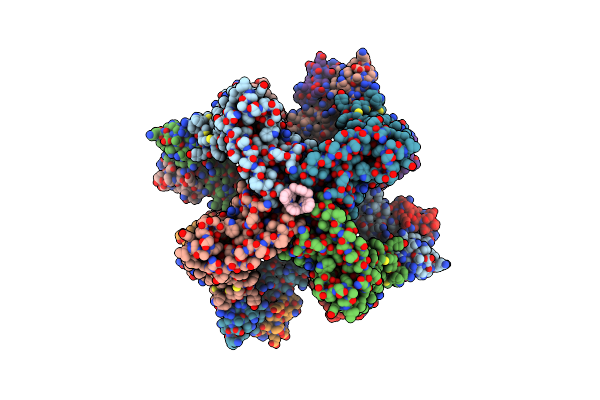 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, Y7Z, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN |
 |
Plasmodium Falciparum Niemann-Pick Type C1-Related Protein Bound With Mmv009108
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, Y6F |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: CLR, NAG |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: CLR, NAG |
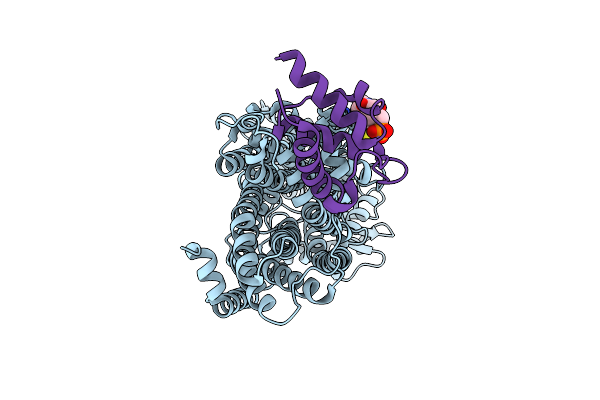 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: TRANSPORT PROTEIN Ligands: PNS |
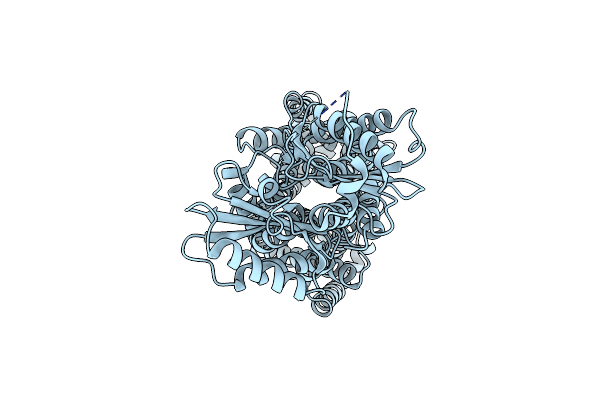 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSPORT PROTEIN |
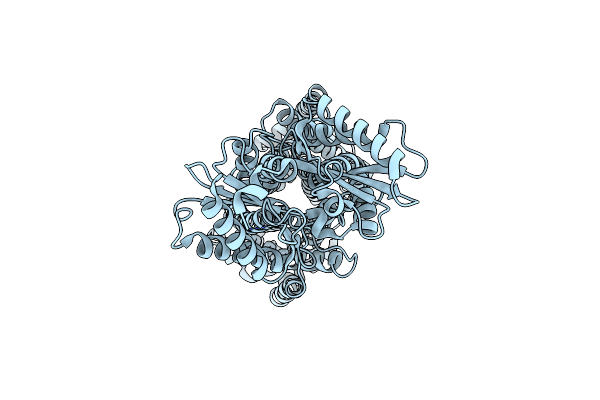 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSPORT PROTEIN |

