Search Count: 183
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-5 (Lro-C2-5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Acidic Ph (Ph 4.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Basic Ph (Ph 8.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C3-7 (Lro-C3-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C4-13 (Lro-C4-13)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C5-1 (Lro-C5-1)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 2 (Lrd-2)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 7 (Lrd-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
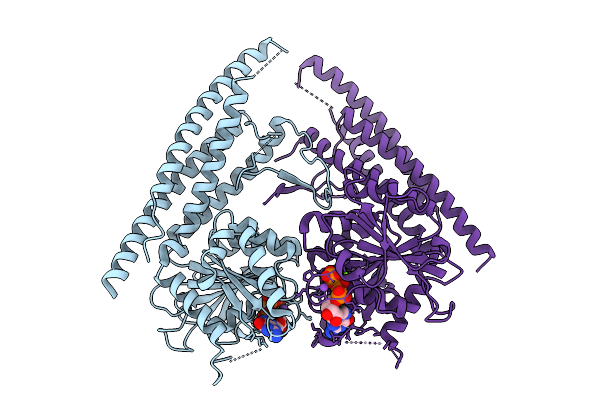 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, NA |
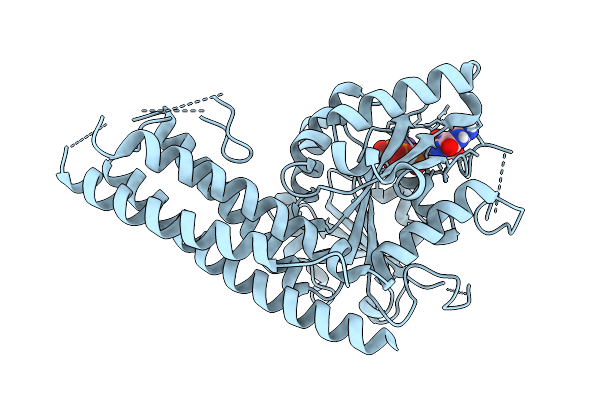 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, K |
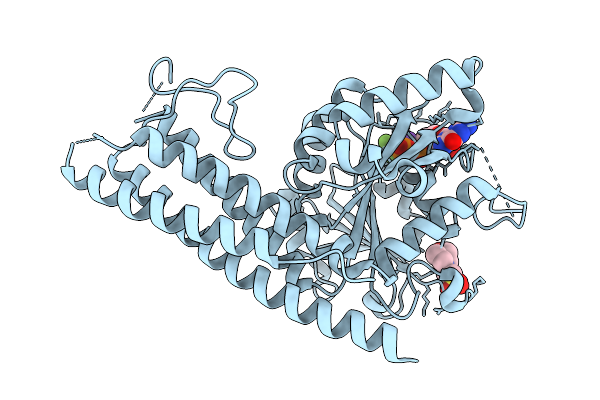 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GDP, BEF, MG, K, MES |
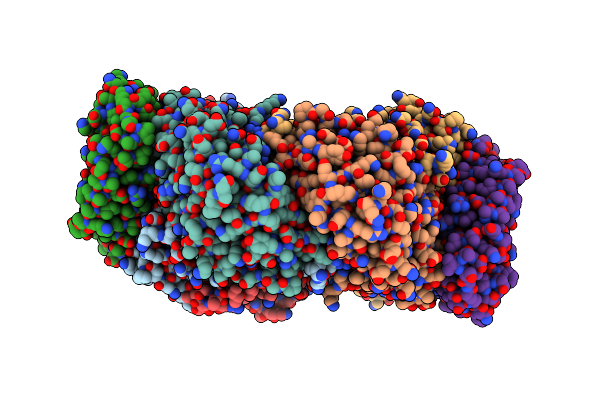 |
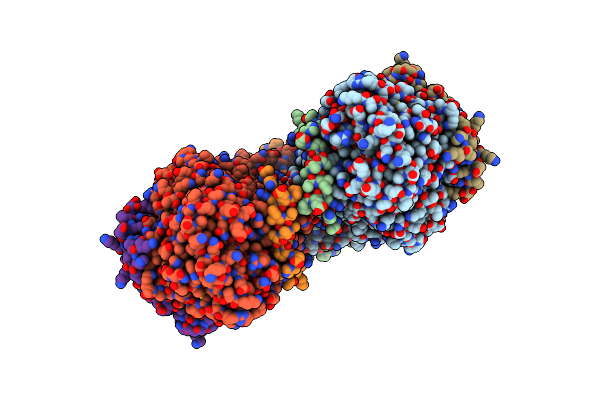
Cryo-Em Complex Structure Between Hydroxylase And Regulatory Component From Soluble Methane Monooxygenase
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
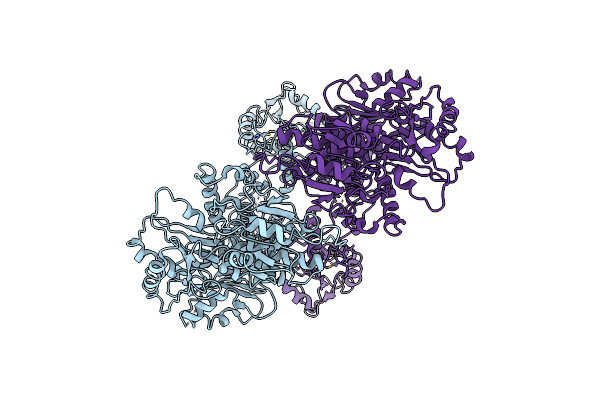
Cryo-Em Structure Of Hydroxylase In Soluble Methane Monooxygenase From Methylosinus Sporium 5
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: ZN |
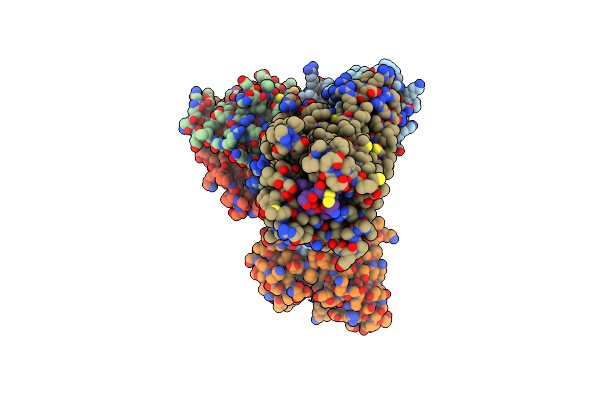 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN |
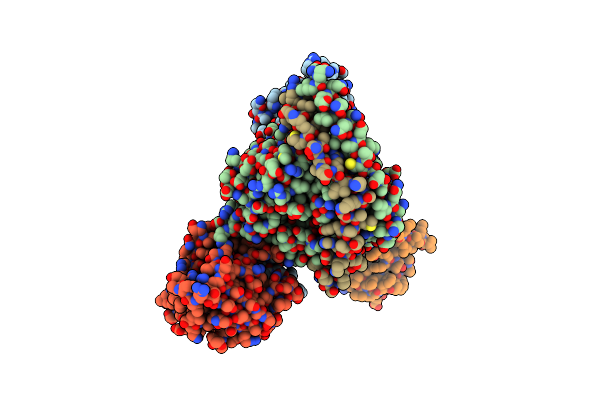 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, ALF, MG |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, MG |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, LMR |

