Search Count: 47
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gmppnp And Covalently Bound To An Adduct Of {(2S)-4-[7-(8-Chloronaphthalen-1-Yl)-2-{[(2S)-1-Methylpyrrolidin-2-Yl]Methoxy}-5,6,7,8-Tetrahydropyrido[3,4-D]Pyrimidin-4-Yl]-1-[(2Z)-2-Fluoro-3-(Pyridin-2-Yl)Prop-2-Enoyl]Piperazin-2-Yl}Acetonitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, A1B7P, MG |
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gdp And Covalently Bound To An Adduct Of (2S)-1-{4-[(7P)-7-(8-Ethynyl-7-Fluoro-3-Hydroxynaphthalen-1-Yl)-8-Fluoro-2-{[(2R,4R,7As)-2-Fluorotetrahydro-1H-Pyrrolizin-7A(5H)-Yl]Methoxy}Pyrido[4,3-D]Pyrimidin-4-Yl]Piperazin-1-Yl}-2-Fluoro-3-(1,3-Thiazol-2-Yl)Propan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1B7Q, MG |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |
 |
Crystal Structure Of Rsegfp2 Mutant V151A In The Non-Fluorescent Off-State Determined By Synchrotron Radiation At 100K
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Rsegfp2 Mutant V151A In The Fluorescent On-State Determined By Synchrotron Radiation At 100K
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 Mutant V151L In The Fluorescent On-State Determined By Synchrotron Radiation At 100K
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 Mutant V151L In The Non-Fluorescent Off-State Determined By Synchrotron Radiation At 100K
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 In The Non-Fluorescent Off-State Determined By Serial Femtosecond Crystallography At Room Temperature
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 Mutant V151A In The Fluorescent On-State Determined By Serial Femtosecond Crystallography At Room Temperature
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 Mutant V151L In The Non-Fluorescent Off-State The Determined By Serial Femtosecond Crystallography At Room Temperature
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Rsegfp2 Mutant V151A In The Non-Fluorescent Off-State Determined By Serial Femtosecond Crystallography At Room Temperature
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Lysozyme Structure Determined From Sfx Data Using A Sheet-On-Sheet Chipless Chip
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2018-10-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-17 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
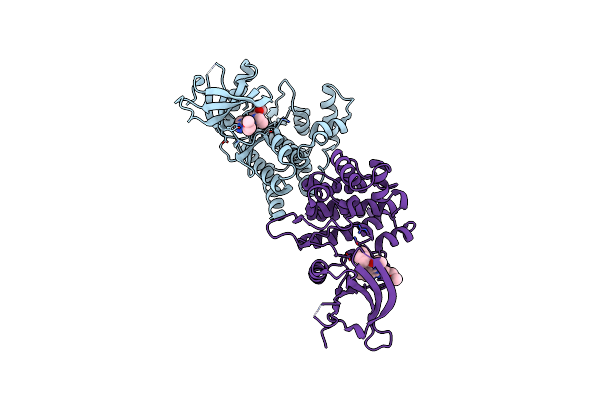 |
Crystal Structure Of Janus Kinase 2 In Complex With N,N-Dicyclopropyl-10-Ethyl-7-[(3-Methoxypropyl)Amino] -3-Methyl-3,5,8,10-Tetraazatricyclo[7.3.0.0,6] Dodeca-1(9),2(6),4,7,11-Pentaene-11-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2015-08-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 50Y |
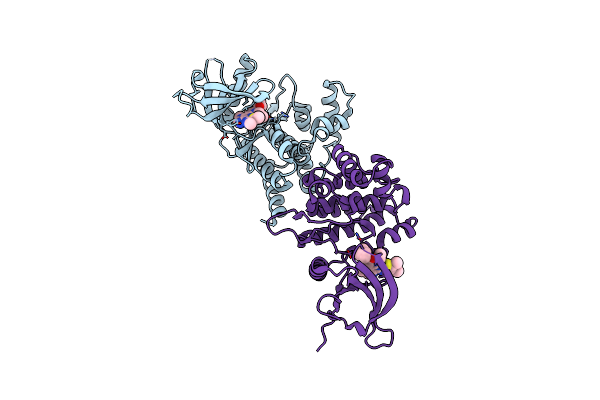 |
Crystal Structure Of Janus Kinase 2 In Complex With N,N-Dicyclopropyl-7-[(Dimethyl-1,3-Thiazol-2-Yl)Amino]-10-Ethyl-3-Methyl-3,5,8,10-Tetraazatricyclo[7.3.0.02,6] Dodeca-1(9),2(6),4,7,11-Pentaene-11-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-08-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 50W |
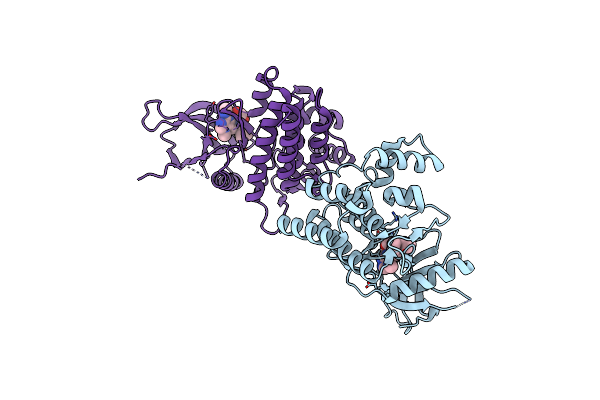 |
Crystal Structure Of Janus Kinase 2 In Complex With N,N-Dicyclopropyl-10-[(2S)-2,3-Dihydroxypropyl]-3-Methyl-7-(Methylamino)-3,5,8,10-Tetraazatricyclo [7.3.0.02,6]Dodeca-1(9),2(6),4,7,11-Pentaene-11-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-08-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 50O |
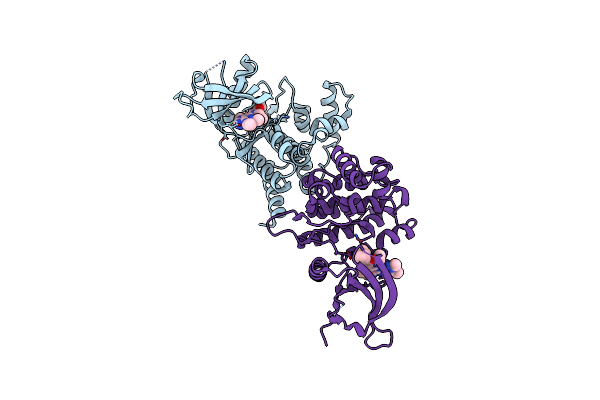 |
Crystal Structure Of Janus Kinase 2 In Complex With N,N-Dicyclopropyl-10-Ethyl-7-[(3-Methoxypropyl)Amino] -3-Methyl-3,5,8,10-Tetraazatricyclo[7.3.0.0,6] Dodeca-1(9),2(6),4,7,11-Pentaene-11-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-08-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 50V |
 |
Crystal Structure Of Janus Kinase 2 In Complex With A 9H-Carbazole-1-Carboxamide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-06-03 Classification: Transferase/Transferase Inhibitor Ligands: 4OK |
 |
Crystal Structure Of Glycerophosphodiester Phosphodiesterase From Thermococcus Kodakarensis Kod1
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: MG |

