Search Count: 29
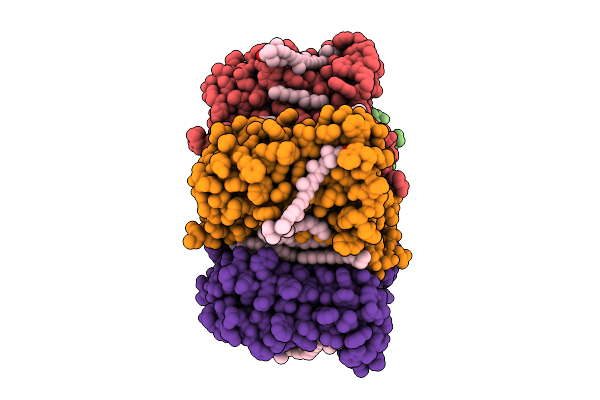 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
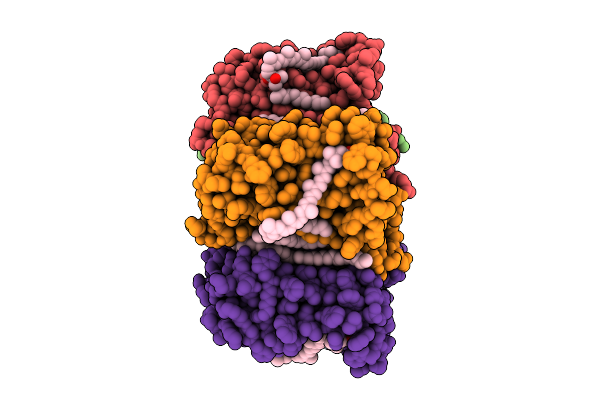 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
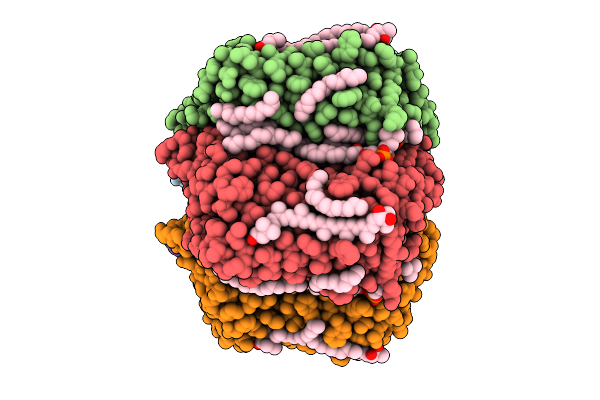 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
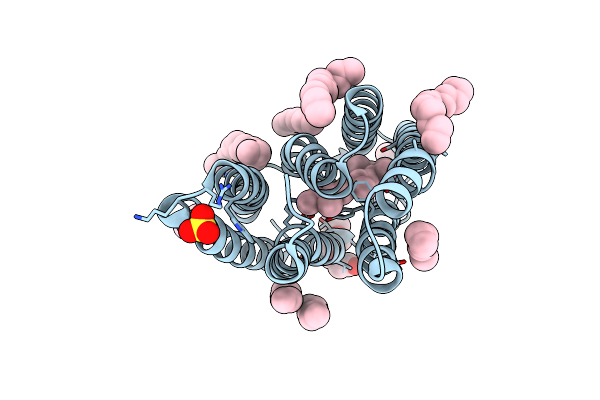 |
The Crystal Structure Of Cyanorhodopsin-Ii (Cyr-Ii) P7104R From Nodosilinea Nodulosa Pcc 7104
Organism: Nodosilinea nodulosa pcc 7104
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: RET, PG4, HEX, OCT, C14, R16, SO4, CL |
 |
Organism: Uncultured bdellovibrionales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: PROTON TRANSPORT Ligands: RET, K3I |
 |
Organism: Bdellovibrio
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3: Resting State Structure With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3 : Structure Obtained 1 Msec After Photoexcitation With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
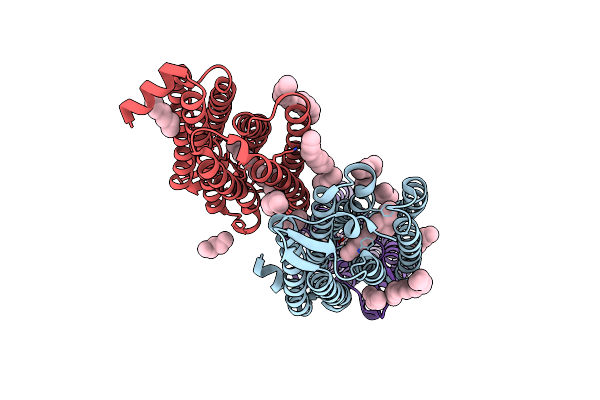 |
Anion Free Form Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3, Structure Determined By Serial Femtosecond Crystallography At Sacla
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, DD9, C14, R16, OCT, CL |
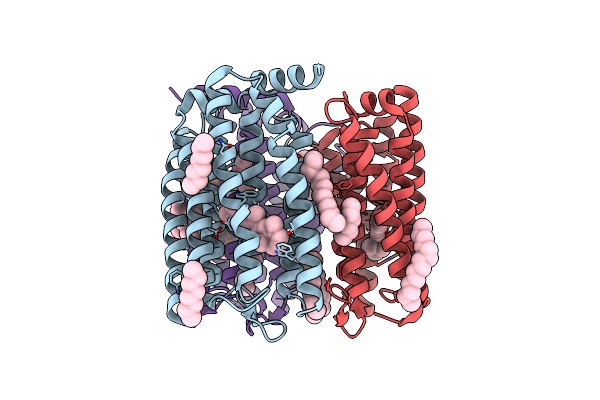 |
The Crystal Structure Of Cyanorhodopsin (Cyr) N2098R From Cyanobacteria Calothrix Sp. Nies-2098
Organism: Calothrix sp. nies-2098
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-10-21 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, OCT, C14, D10 |
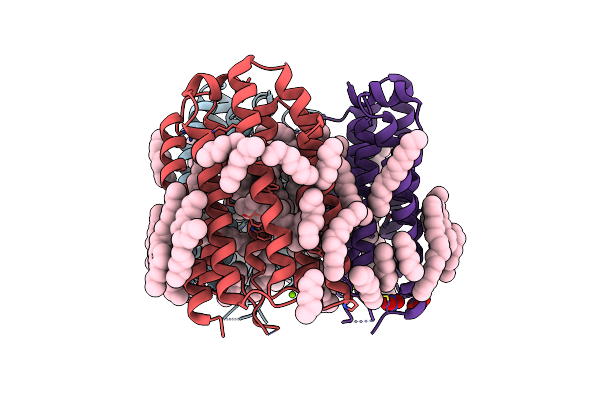 |
The Crystal Structure Of Cyanorhodopsin (Cyr) N4075R From Cyanobacteria Tolypothrix Sp. Nies-4075
Organism: Tolypothrix sp. nies-4075
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-10-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG, HEX, OCT, D10, D12, R16, C14, NO3 |
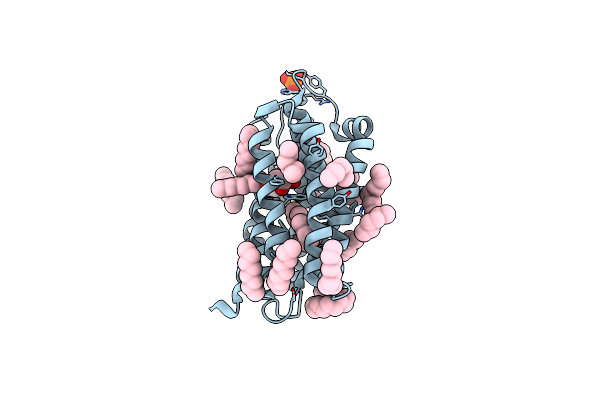 |
Crystal Structure Of The Dts-Motif Rhodopsin From Phaeocystis Globosa Virus 12T
Organism: Phaeocystis globosa virus 12t
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-10-02 Classification: MEMBRANE PROTEIN Ligands: RET, OCT, PO4, HEX, D10, D12, DD9, OLB |
 |
Crystal Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Clp, From Nonlabens Marinus
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2016-07-13 Classification: TRANSPORT PROTEIN Ligands: RET, CL, HEX, OCT, D12, D10 |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: MEMBRANE PROTEIN Ligands: RET, OLA, PEG |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
 |
Structure Of The Mrna Binding Fragment Of Elongation Factor Selb In Complex With Secis Rna.
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-11-06 Classification: translation/RNA Ligands: MG, NA, CA, CL |
 |
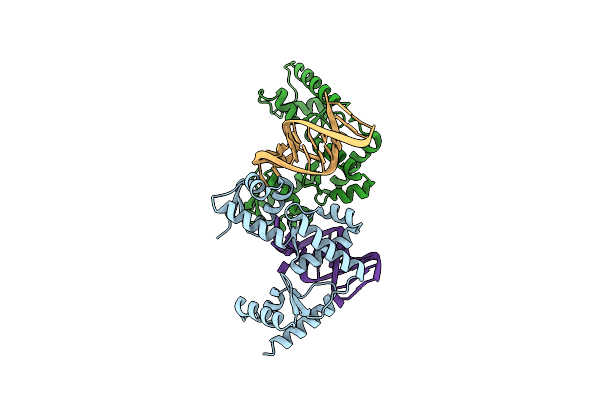
Structure Of The Mrna-Binding Domain Of Elongation Factor Selb From E.Coli In Complex With Secis Rna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-30 Classification: translation/RNA Ligands: MG, CL, CA, NA |
 |
C-Terminal Domain(Wh2-Wh4) Of Elongation Factor Selb In Complex With Secis Rna
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2007-05-08 Classification: TRANSLATION |
 |

