Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
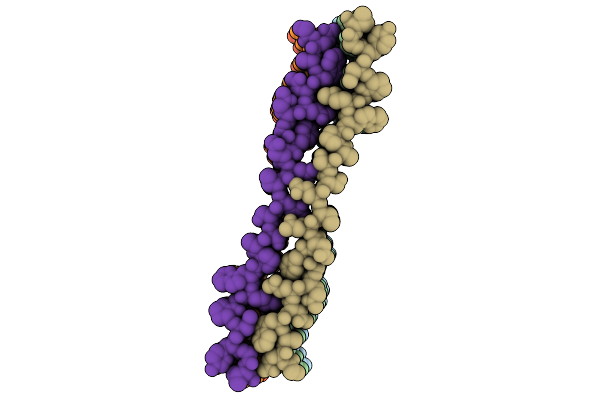 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
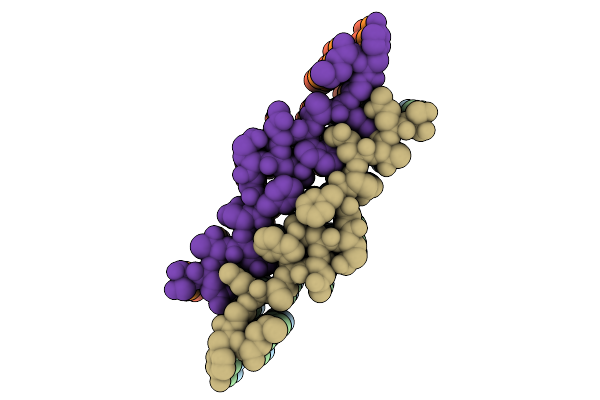 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
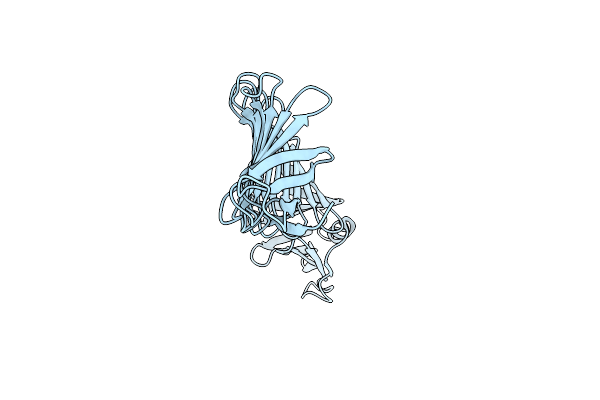 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-27 Classification: UNKNOWN FUNCTION |
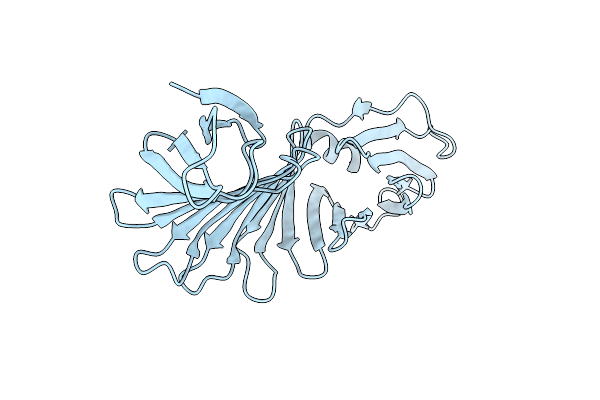 |
Ospa Mutant, Psam-Vlgdv1-Form3, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Ospa Mutant, Psam-Vlgdv1-Form4, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Ospa Mutant, Psam-Vlgdv1-Form5, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Ospa Mutant, Psam-Vlgdv1-Form6, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Ospa Mutant, Psam-Vlgdv1-Form7, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Ospa Mutant, Psam-Vlgdv1-Form8, Grafted Short Chameleon Sequence From Alpha-B Crystallin
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Organism: Borrelia burgdorferi (strain atcc 35210 / b31 / cip 102532 / dsm 4680)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Organism: Borrelia burgdorferi (strain atcc 35210 / b31 / cip 102532 / dsm 4680)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Organism: Borrelia burgdorferi (strain atcc 35210 / b31 / cip 102532 / dsm 4680)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
 |
Organism: Borrelia burgdorferi (strain atcc 35210 / b31 / cip 102532 / dsm 4680)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-27 Classification: LIPID BINDING PROTEIN |
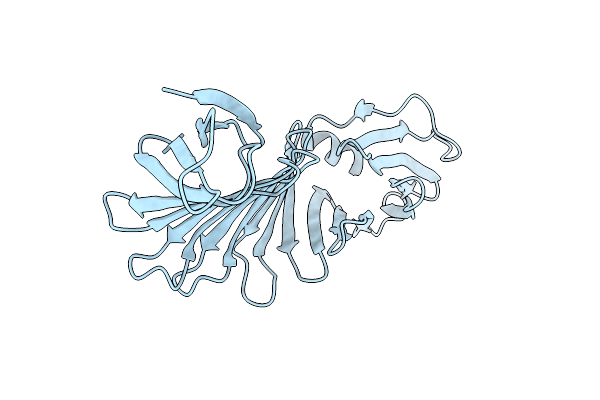 |
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-18 Classification: MEMBRANE PROTEIN |
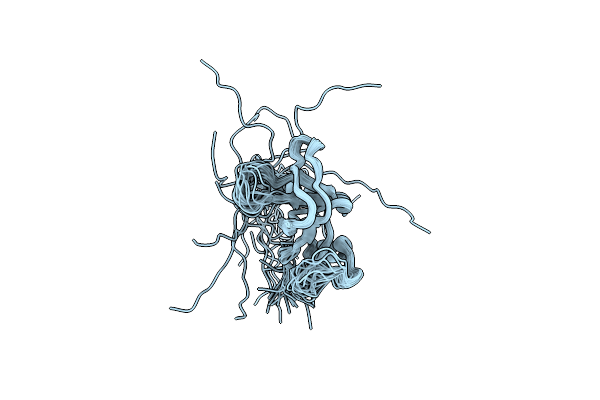 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2010-04-21 Classification: SIGNALING PROTEIN |
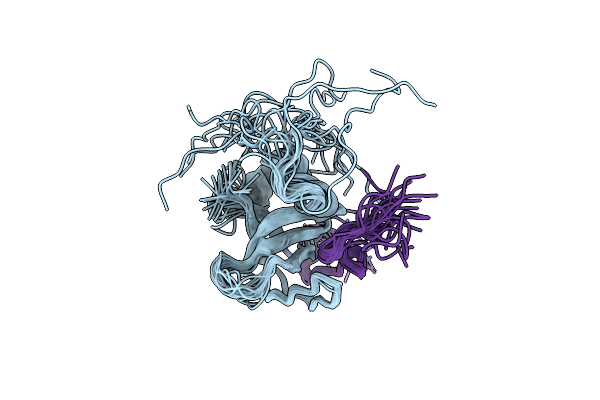 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2010-04-21 Classification: SIGNALING PROTEIN |
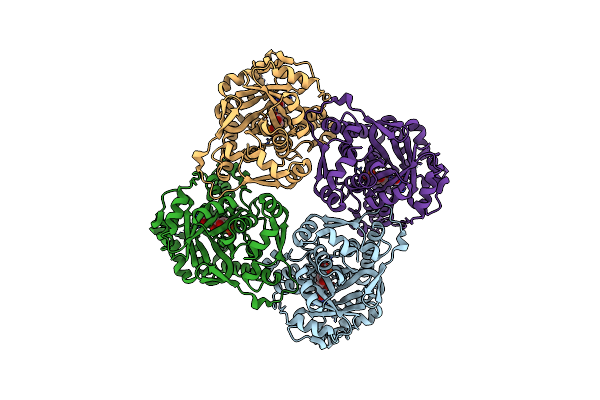 |
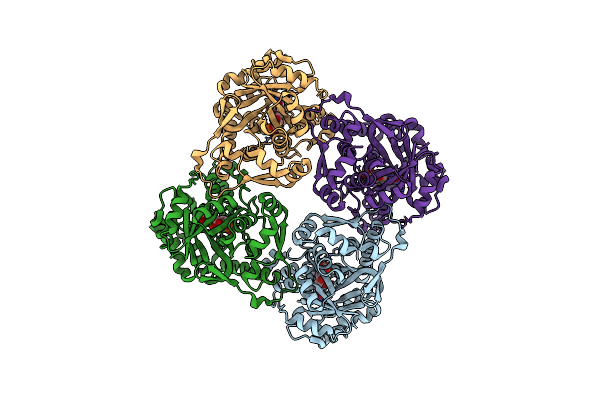
Crystal Structure Of Sulfolobus Shibatae Isopentenyl Diphosphate Isomerase In Complex With Fmn
Organism: Sulfolobus shibatae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-01-20 Classification: ISOMERASE Ligands: FMN |
 |
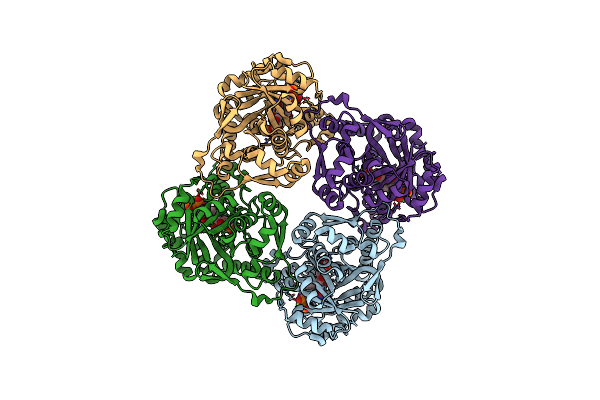
Crystal Structure Of Sulfolobus Shibatae Isopentenyl Diphosphate Isomerase In Complex With Reduced Fmn.
Organism: Sulfolobus shibatae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-01-20 Classification: ISOMERASE Ligands: FNR |
 |
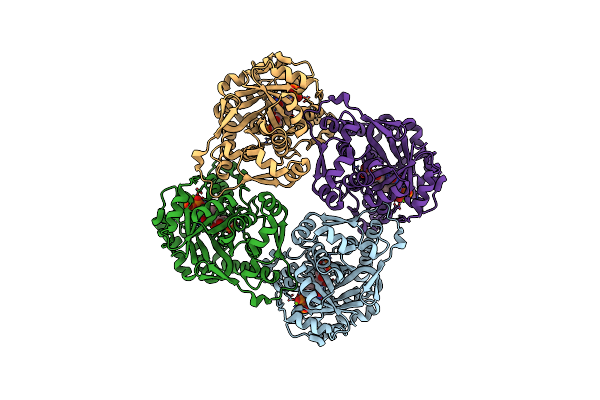
Crystal Structure Of Sulfolobus Shibatae Isopentenyl Diphosphate Isomerase In Complex With Fmn And Ipp.
Organism: Sulfolobus shibatae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-01-20 Classification: ISOMERASE Ligands: FMN, IPR, MG |
 |
Crystal Structure Of Sulfolobus Shibatae Isopentenyl Diphosphate Isomerase In Complex With Fmn And Dmapp.
Organism: Sulfolobus shibatae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-01-20 Classification: ISOMERASE Ligands: FMN, DMA, MG |