Search Count: 40
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: I5G, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: PG4, PGE, N39, N2U |
 |
Organism: Chaetoceros tenuissimus dna virus type-ii
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: VIRUS |
 |
Structure Of Tim-3 In Complex With 8-Chloro-2-Methyl-9-(3-Mehtylpyridin-4-Yl)-[1,2,4]Triazolo[1,5-C]Quinazolin-5(6H)-One (Compound 22)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-10-13 Classification: IMMUNE SYSTEM Ligands: YQ7, CA |
 |
Structure Of Tim-3 In Complex With N-(4-(8-Chloro-2-Mehtyl-5-Oxo-5,6-Dihydro-[1,2,4]Triazolo[1,5-C]Quinazolin-9-Yl)-3-Methylphenyl)Methanesulfonamdide (Compound 35)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-10-13 Classification: IMMUNE SYSTEM Ligands: YQD, CA |
 |
Structure Of Tim-3 In Complex With N-(4-(8-Chloro-2-Methyl-5-Oxo-5,6-Dihydro-[1,2,4]Traizolo[1,5-C]Quinazolin-9-Yl)-3-Methylphenyl)-1H-Imidazole-2-Sulfonamide (Compound 38)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-10-13 Classification: IMMUNE SYSTEM Ligands: YQG, CA |
 |
Crystal Structure Of Human P-Cadherin Mec12 (X Dimer) In Complex With 2-(5-Chloro-2-Methyl-1H-Indol-3-Yl)Ethan-1-Amine (Inhibitor)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-09-15 Classification: CELL ADHESION Ligands: DMS, G60, CA |
 |
Crystal Structure Of Human P-Cadherin Rec12 (Monomer) In Complex With 2-(5-Chloro-2-Methyl-1H-Indol-3-Yl)Ethan-1-Amine (Inhibitor)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-09-15 Classification: CELL ADHESION Ligands: CA, G60 |
 |
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: SO4 |
 |
Cholesterol Esterase From Burkholderia Stabilis (Orthorhombic Crystal Form)
Organism: Burkholderia stabilis
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-12-16 Classification: HYDROLASE Ligands: CA, GOL, IMD, IPA |
 |
Organism: Burkholderia stabilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-16 Classification: HYDROLASE Ligands: CA |
 |
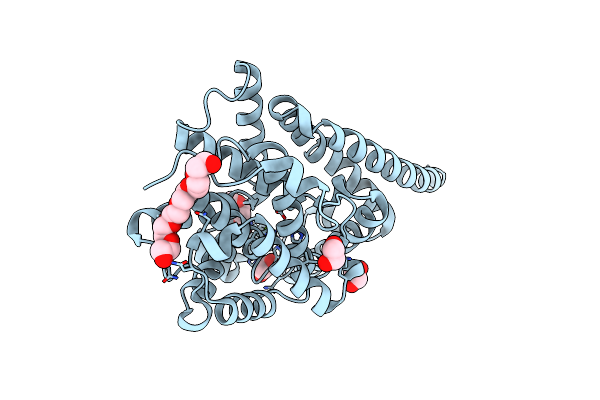
Crystal Structure Of The Transmembrane Domain Of Salpingoeca Rosetta Rhodopsin Phosphodiesterase
Organism: Salpingoeca rosetta (strain atcc 50818 / bsb-021)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
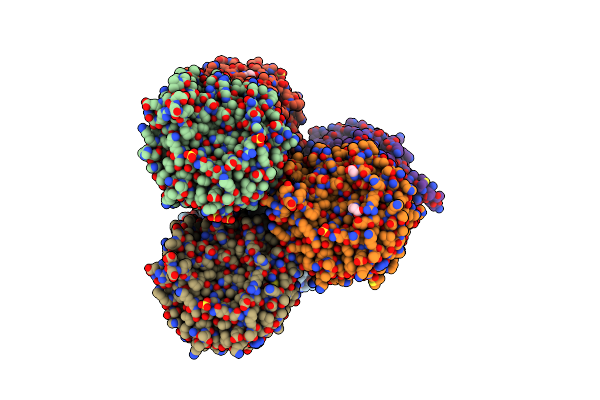
Crystal Structure Of The Phosphodiesterase Domain Of Salpingoeca Rosetta Rhodopsin Phosphodiesterase
Organism: Salpingoeca rosetta (strain atcc 50818 / bsb-021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN Ligands: 2PE, PEG, EDO, ZN, MG |
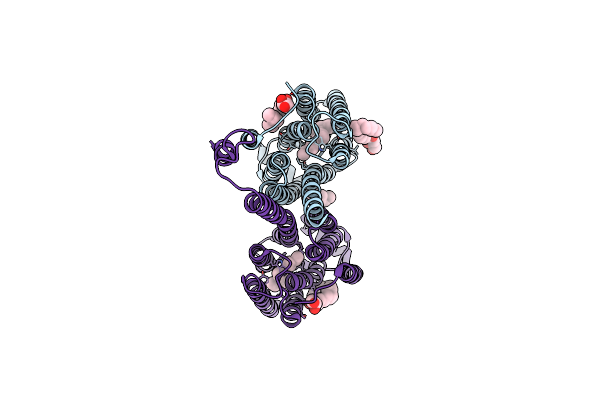 |
Crystal Structure Of The Transmembrane Domain And Linker Region Of Salpingoeca Rosetta Rhodopsin Phosphodiesterase
Organism: Salpingoeca rosetta (strain atcc 50818 / bsb-021)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: SO4, BAL, GLY |
 |
Crystal Structure Of Ferredoxin: Thioredoxin Reductase And Thioredoxin Y1 Complex
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2020-10-14 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
Crystal Structure Of Ferredoxin: Thioredoxin Reductase And Thioredoxin F2 Complex
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-10-14 Classification: ELECTRON TRANSPORT Ligands: SF4, NO3, MG |
 |
Crystal Structure Of Ferredoxin: Thioredoxin Reductase And Thioredoxin M2 Complex
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-10-14 Classification: ELECTRON TRANSPORT Ligands: SF4, NA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2020-10-14 Classification: ELECTRON TRANSPORT |
 |
2'-Fucosyllactose And 3-Fucosyllactose Binding Protein From Bifidobacterium Longum Infantis, Bound With 2'-Fucosyllactose
Organism: Bifidobacterium longum subsp. infantis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-09-04 Classification: TRANSPORT PROTEIN Ligands: PG0, BGC, ZN, MES |

