Search Count: 35
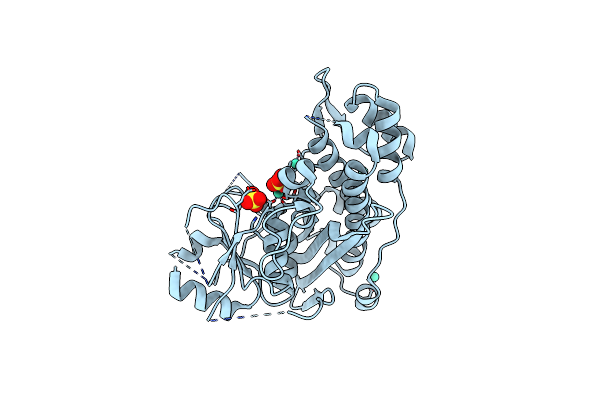 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase Inpp1 In Complex Gadolinium After Addition Of Inositol 1,3,4-Trisphosphate At 2.5 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-11-25 Classification: SIGNALING PROTEIN Ligands: SO4, GD |
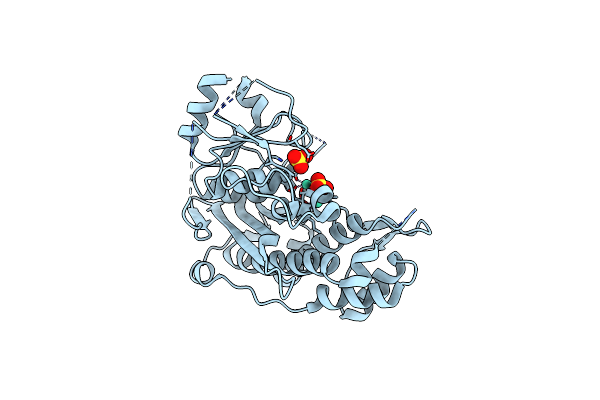 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase Inpp1 In Complex Gadolinium After Addition Of Inositol 1,3,4-Trisphosphate And Lithium At 3.2 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-11-25 Classification: SIGNALING PROTEIN Ligands: GD, SO4 |
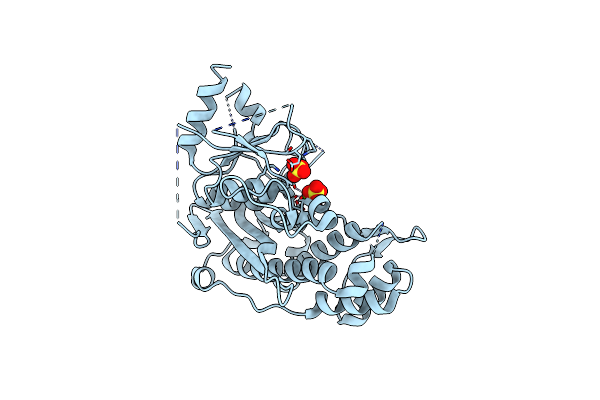 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase (Inpp1) D54A Mutant
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: CA, SO4 |
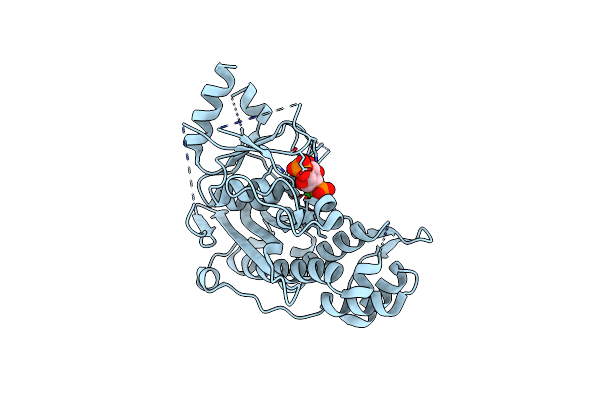 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase (Inpp1) D54A Mutant In Complex With Inositol (1,4)-Bisphosphate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-25 Classification: HYDROLASE Ligands: 2IP, CA |
 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase Inpp1 In Complex Gadolinium But No Lithium At 3 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-18 Classification: SIGNALING PROTEIN Ligands: GD, SO4 |
 |
Crystal Structure Of Inositol Polyphosphate 1-Phosphatase Inpp1 In Complex Gadolinium And Lithium At 2.5 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-18 Classification: SIGNALING PROTEIN Ligands: GD, SO4 |
 |
Crystal Structure Of Human Diphosphoinositol Polyphosphate Phosphohydrolase 1 In Complex With 1-Ip7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: O81, MG, CL |
 |
Crystal Structure Of Human Diphosphoinositol Polyphosphate Phosphohydrolase 1 In Complex With 5-Ip7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: I7P, MG, CL |
 |
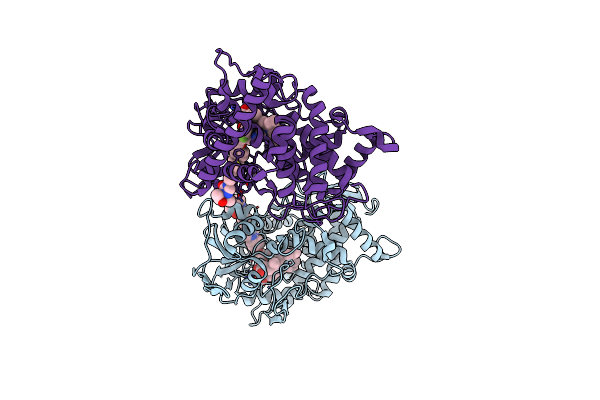
Direct Activation Of The Executioner Domain Of Mlkl By A Select Repertoire Of Inositol Phosphates
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-05-15 Classification: LIPID BINDING PROTEIN |
 |
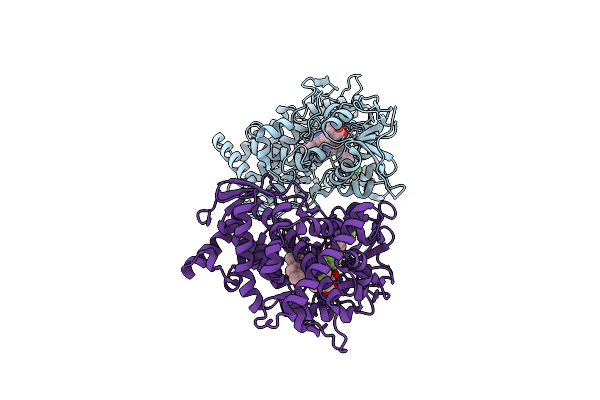
Crystal Structure Of Sterol 14-Alpha Demethylase (Cyp51) From A Pathogenic Yeast Candida Albicans In Complex With The Antifungal Drug Posaconazole
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: HEM, X2N |
 |
Crystal Structure Of Sterol 14-Alpha Demethylase (Cyp51) From Candida Albicans In Complex With The Tetrazole-Based Antifungal Drug Candidate Vt1161 (Vt1)
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, VT1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-03-01 Classification: MEMBRANE PROTEIN Ligands: 755, GSH, BOG, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-03-01 Classification: MEMBRANE PROTEIN Ligands: 758, GSH, BOG, PG4 |
 |
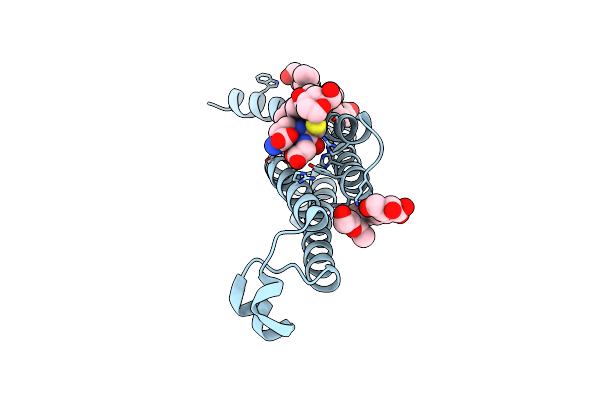
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-03-01 Classification: Membrane Protein, Transferase Ligands: 7DN, GSH, JZR, BOG, PG4, PEG |
 |
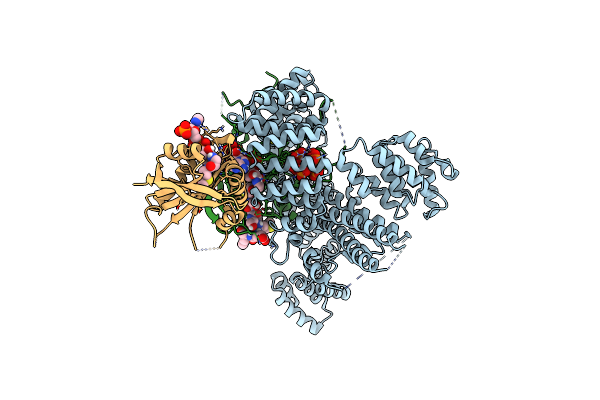
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-09-14 Classification: Isomerase/Isomerase Inhibitor Ligands: 6PW, GSH, BOG, PG4 |
 |
Crystal Structure Of Yeast N-Terminal Acetyltransferase Nate (Ip6) In Complex With A Bisubstrate
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-07-20 Classification: TRANSFERASE Ligands: IHP, ACO, CMC |
 |
Crystal Structure Of Yeast N-Terminal Acetyltransferase Nate (Ppgpp) In Complex With A Bisubstrate
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-07-20 Classification: TRANSFERASE Ligands: G4P, ACO, CMC |
 |
Crystal Structure Of Yeast N-Terminal Acetyltransferase (Ppgpp) Nate In Complex With A Bisubstrate
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2016-07-13 Classification: TRANSFERASE Ligands: G4P, CMC, ACO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2016-04-13 Classification: Isomerase/Isomerase inhibitor Ligands: 4UJ, GSH, JZR, BOG, PGE, PEG |
 |
Discovery Of A Potent And Selective Mpges-1 Inhibitor For The Treatment Of Pain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-04-13 Classification: Isomerase/isomerase inhibitor Ligands: 4UK, GSH, BOG, PG4, 1KA |

