Search Count: 36
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: Q35 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: T35, MYR, GOL |
 |
The Agonist-Free Structure Of Human Ppargamma Ligand Binding Domain In The Presence Of The Src-1 Coactivator Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-07-05 Classification: TRANSCRIPTION Ligands: MYR, GOL |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-09-02 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2015-09-02 Classification: HYDROLASE Ligands: DAL, GOL |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: ZN, SO4, GOL |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: ZN, NI, PO4, GOL |
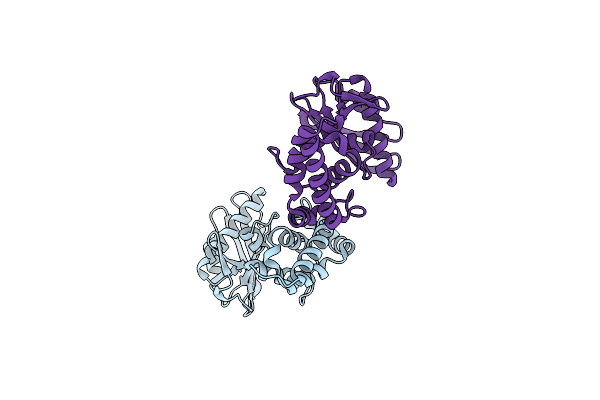 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-04-09 Classification: TRANSFERASE |
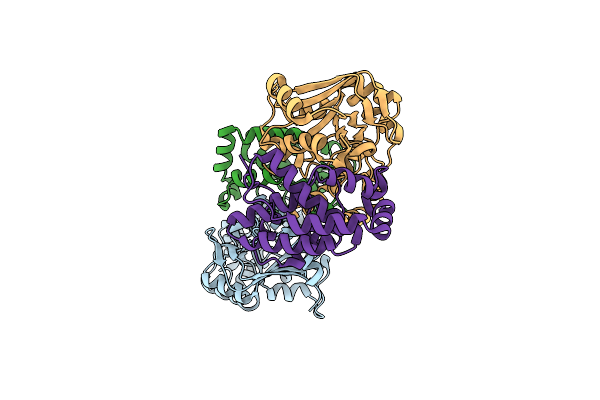 |
High-Resolution Crystal Structure Streptococcus Pyogenes Beta-Nad+ Glycohydrolase In Complex With Its Endogenous Inhibitor Ifs Reveals A Water-Rich Interface
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2013-10-30 Classification: Hydrolase/Hydrolase Inhibitor |
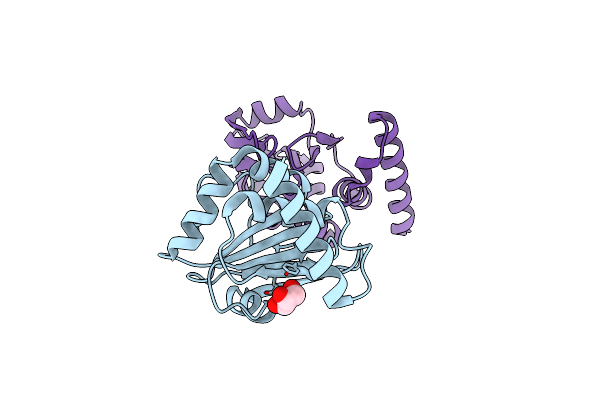 |
Structural And Functional Characterizations Of A Thioredoxin-Fold Protein From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: GOL |
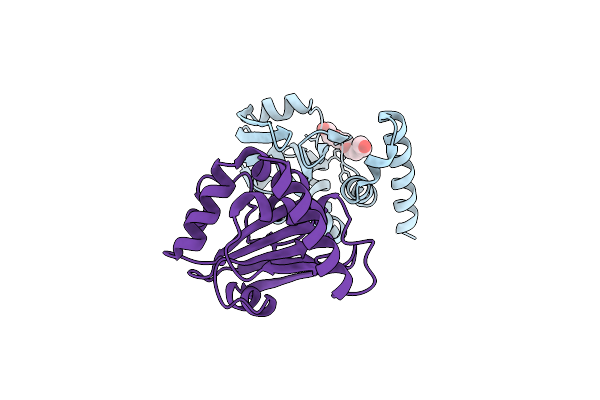 |
Structural And Functional Characterizations Of A Thioredoxin-Fold Protein From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: PG4 |
 |
Structural Basis For The Inhibition Of Mycobacterium Tuberculosis L,D-Transpeptidase By Meropenem, A Drug Effective Against Extensively Drug-Resistant Strains
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-02-27 Classification: TRANSFERASE Ligands: CA, GOL |
 |
Structural Basis For The Inhibition Of Mycobacterium Tuberculosis L,D-Transpeptidase By Meropenem, A Drug Effective Against Extensively Drug-Resistant Strains
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2013-02-27 Classification: TRANSFERASE Ligands: EMT, GOL |
 |
Structural Basis For The Inhibition Of Mycobacterium Tuberculosis L,D-Transpeptidase By Meropenem, A Drug Effective Against Extensively Drug-Resistant Strains
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-02-27 Classification: TRANSFERASE Ligands: DWZ |
 |
Crystal Structure Of Pyridoxal Biosynthesis Lyase Pdxs From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-11-14 Classification: LYASE |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-11-14 Classification: LYASE Ligands: R5P |
 |
Mycobacterium Tuberculosis Eis Protein Initiates Modulation Of Host Immune Responses By Acetylation Of Dusp16/Mkp-7
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-25 Classification: TRANSFERASE |
 |
Crystal Structure Of Enhanced Intracellular Survival (Eis) Protein From Mycobacterium Tuberculosis With Acetyl Coa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-05-23 Classification: TRANSFERASE Ligands: ACO |
 |
Mycobacterium Tuberculosis Eis Protein Initiates Modulation Of Host Immune Responses By Acetylation Of Dusp16/Mkp-7
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2012-05-23 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-23 Classification: TRANSFERASE |

