Search Count: 44
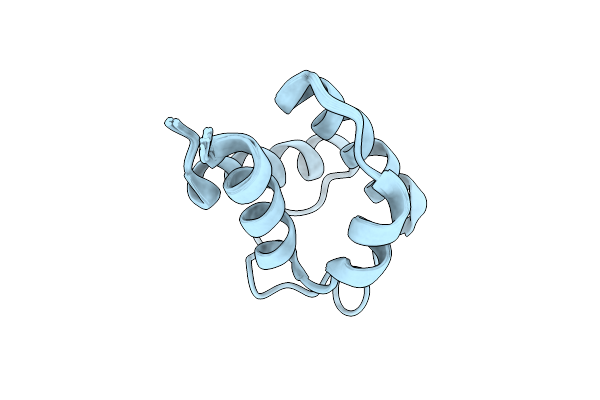 |
Solution Structure Of Holo Acyl Carrier Protein 2 (Apef) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
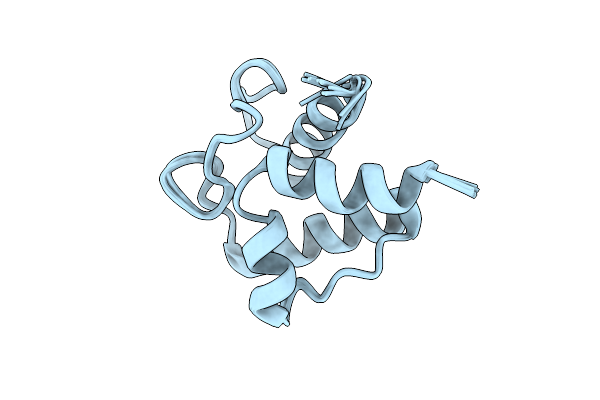 |
Solution Structure Of Holo Acyl Carrier Protein 1 (Apee) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
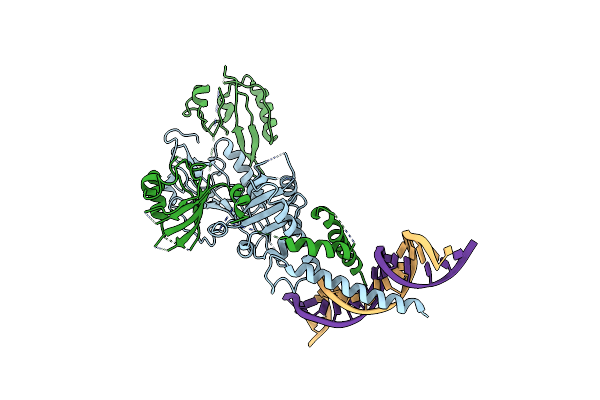 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSCRIPTION/DNA |
 |
Organism: Deinococcus geothermalis (strain dsm 11300 / cip 105573 / ag-3a)
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: CARBOHYDRATE |
 |
Covalently Stabilized Triangular Trimer Derived From Abeta16-36 With P-Iodo-Phenylalanine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-05-31 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-05-31 Classification: DE NOVO PROTEIN Ligands: MPD, CL |
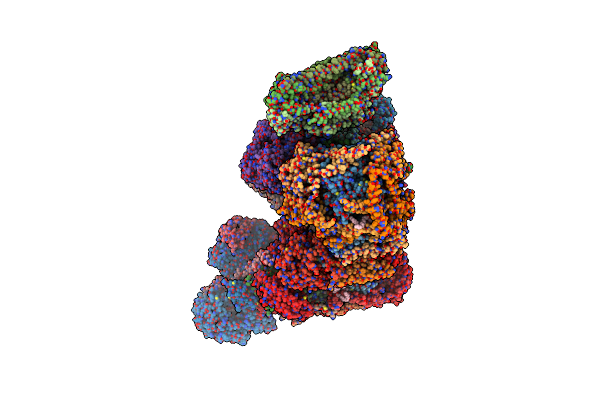 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: PC1, 3PE, FES, CDL, FMN, SF4, CU, HEA, MG, ZN, NAP, HEM, HEC, UQ2 |
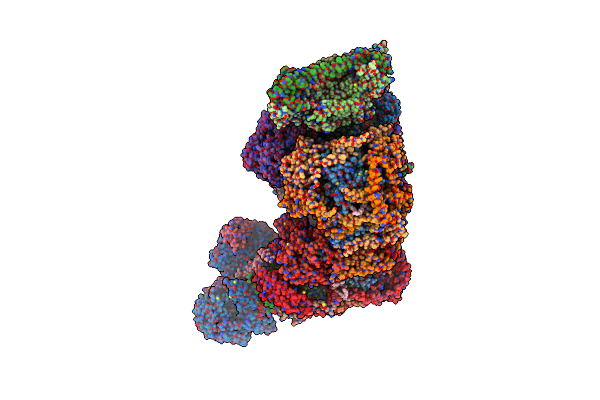 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: FES, 3PE, CDL, PC1, FMN, SF4, ZN, NAP, HEM, HEC, HEA, CU, MG |
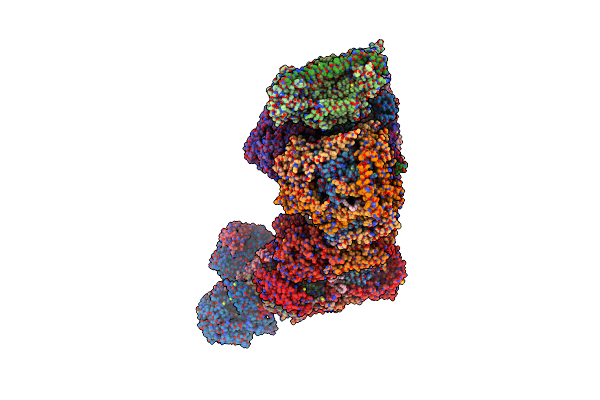 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: FES, CDL, 3PE, PC1, FMN, SF4, ZN, NAP, HEM, HEC, HEA, CU, MG |
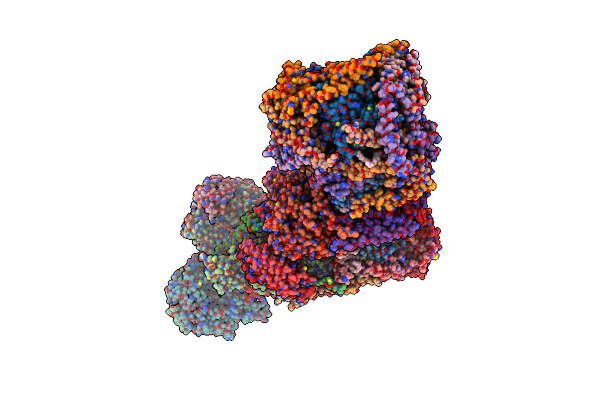 |
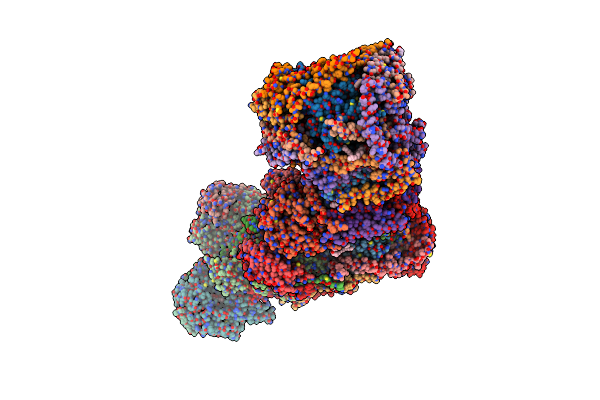
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: 3PE, CDL, FMN, SF4, FES, ZN, PC1, NAP |
 |
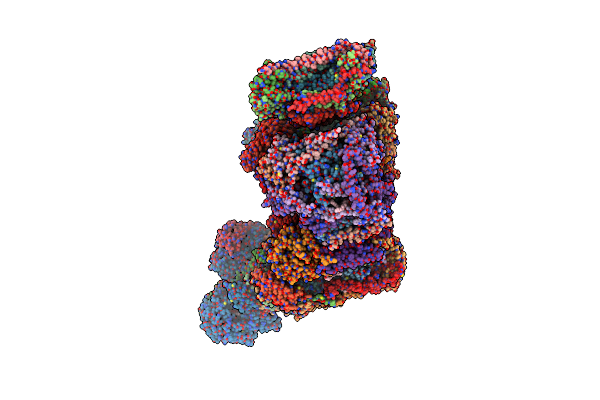
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: 3PE, CDL, FMN, SF4, FES, ZN, NAP, PC1 |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, FES, 3PE, CDL, FMN, SF4, ZN, NAP, PC1, HEA, CU, MG |
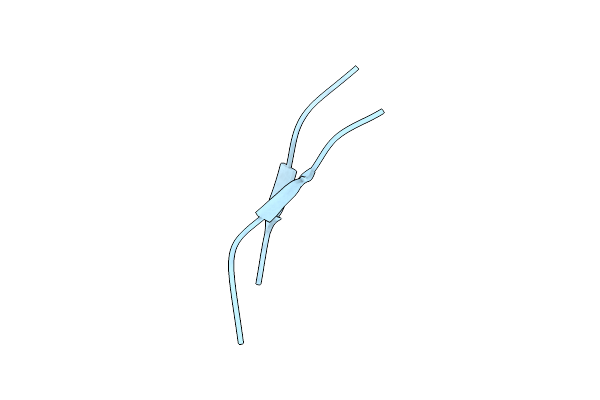 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-09-08 Classification: DE NOVO PROTEIN Ligands: IOD |
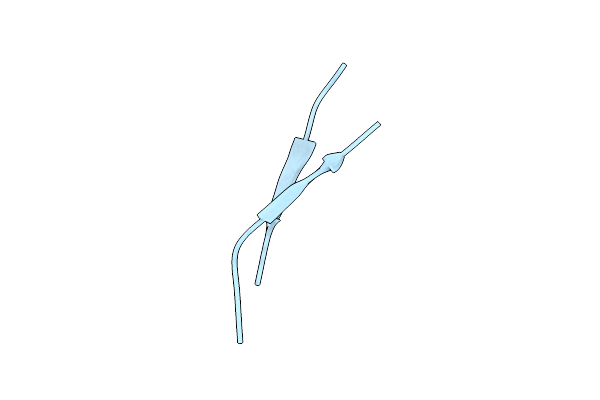 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-09-08 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-09-08 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-09-08 Classification: DE NOVO PROTEIN |
 |
X-Ray Crystallographic Structure Of A Covalent Trimer Derived From A-Beta 17_36 Containing The I31V Point Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-10-17 Classification: DE NOVO PROTEIN Ligands: HEZ |
 |
X-Ray Crystallographic Structure Of A Covalent Trimer Derived From A-Beta 17_36 Containing The I31Chg Point Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2018-10-17 Classification: DE NOVO PROTEIN Ligands: MPD |
 |
X-Ray Crystallographic Structure Of A Covalent Trimer Derived From A-Beta 17_36 Containing The F20Cha Point Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-10-17 Classification: DE NOVO PROTEIN |

