Search Count: 21
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Form A)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structures Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Form B)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structures Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate A)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
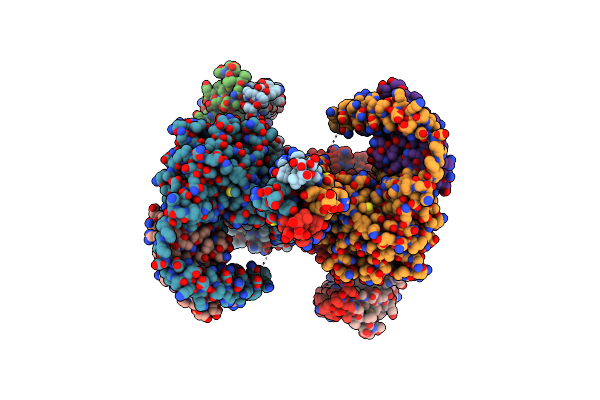 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate B)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate C)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Cryo-Em Structure Of The Head Region Of Full-Length Ergic-53 With Mcfd2 (Substate D)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: PROTEIN TRANSPORT Ligands: CA, ZN |
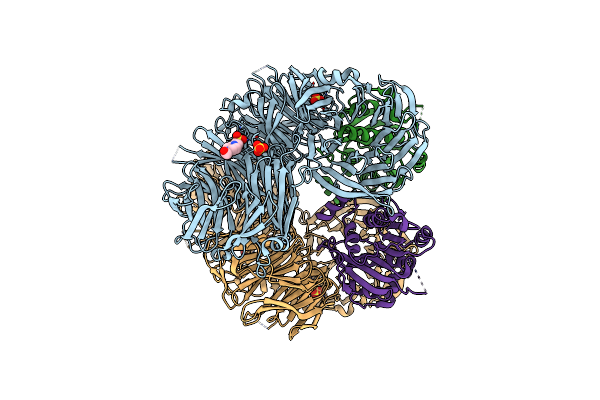 |
The X-Ray Structure Of Yeast Trna Methyltransferase Complex Of Trm7 And Trm734 Essential For 2'-O-Methylation At The First Position Of Anticodon In Specific Trnas
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: SO4, EPE |
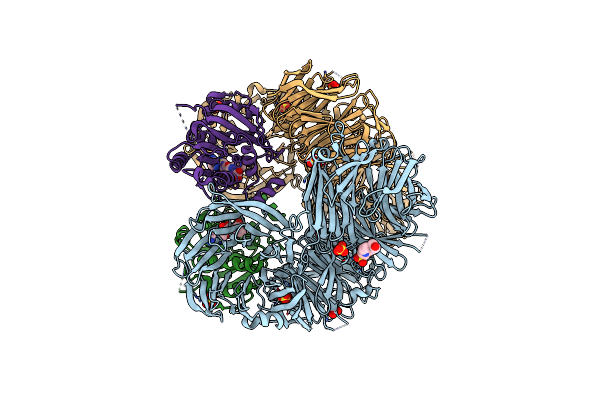 |
The X-Ray Structure Of Yeast Trna Methyltransferase Trm7-Trm734 In Complex With S-Adenosyl-L-Methionine
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: EPE, SO4, SAM |
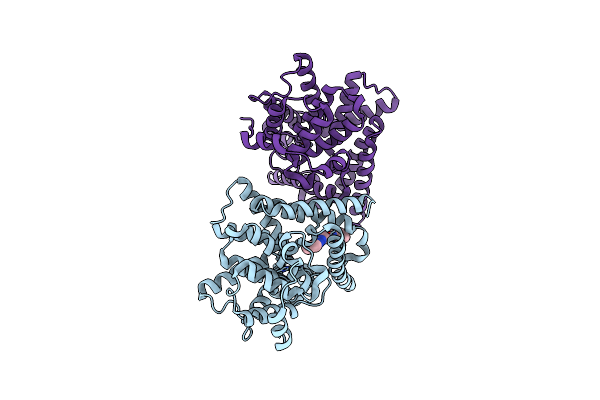 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2019-08-14 Classification: HYDROLASE Ligands: ZN, MG, D6X |
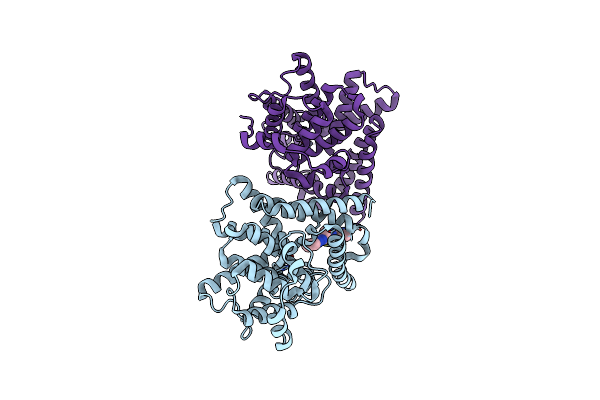 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-08-14 Classification: HYDROLASE Ligands: ZN, MG, D79 |
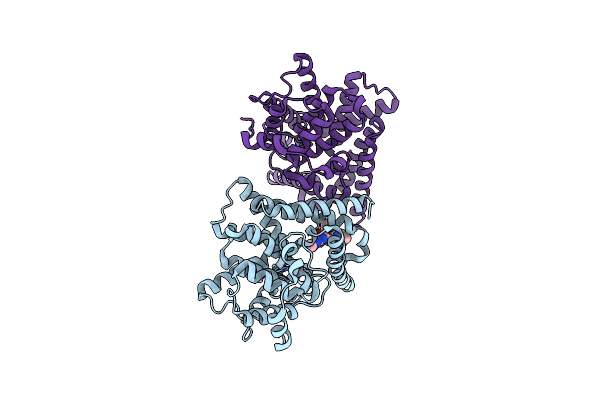 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-08-14 Classification: HYDROLASE Ligands: ZN, MG, D7C |
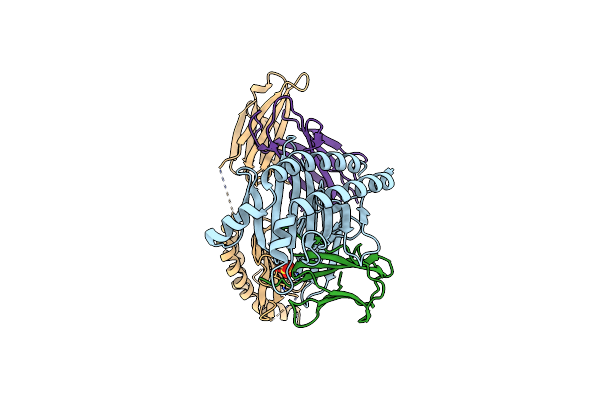 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-12-05 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of The Flexible Tandem Repeat Domain Of Bacterial Cellulose Synthase Subunit C
Organism: Enterobacter sp. cjf-002
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2017-11-01 Classification: BIOSYNTHETIC PROTEIN |
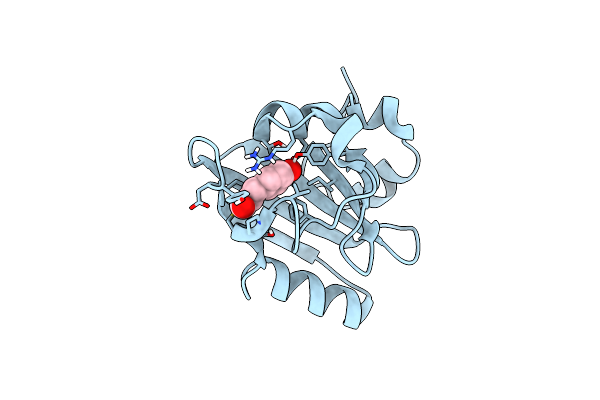 |
Organism: Halorhodospira halophila
Method: NEUTRON DIFFRACTION Resolution:1.49 Å Release Date: 2017-08-30 Classification: SIGNALING PROTEIN Ligands: HC4, DOD |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |

