Search Count: 20
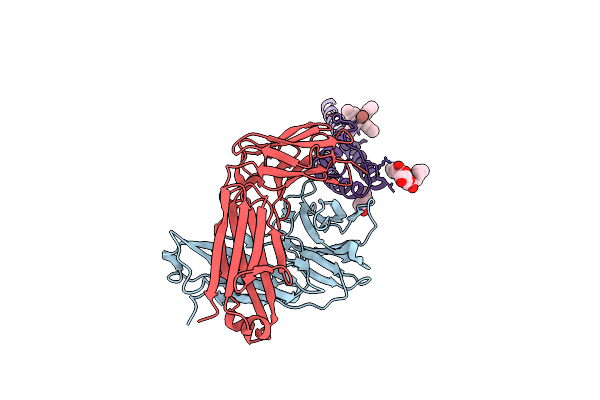 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-02-20 Classification: MEMBRANE PROTEIN Ligands: F09, TL, L2C, TBA |
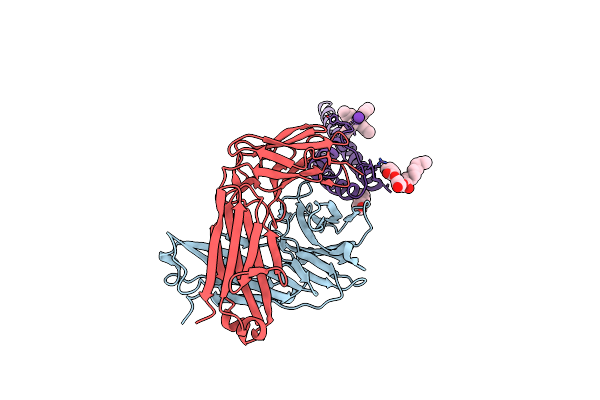 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-02-20 Classification: MEMBRANE PROTEIN Ligands: RB, L2C, F09, TBA |
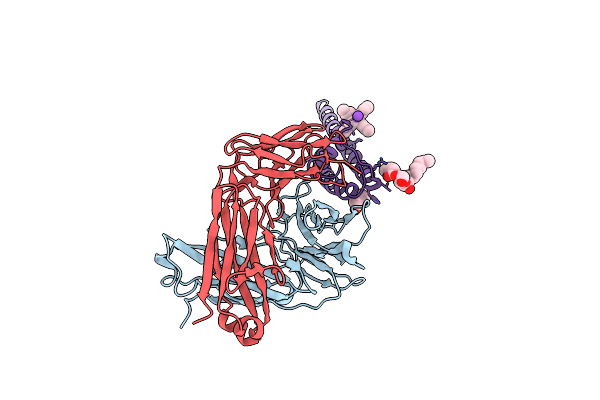 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2007-02-20 Classification: MEMBRANE PROTEIN Ligands: F09, K, L2C, TBA |
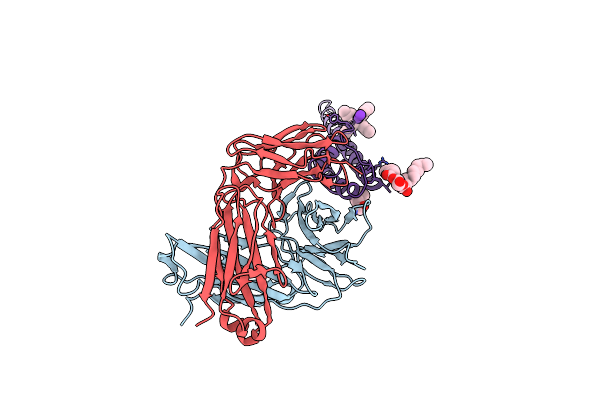 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-02-20 Classification: MEMBRANE PROTEIN Ligands: F09, K, L2C, TBA |
 |
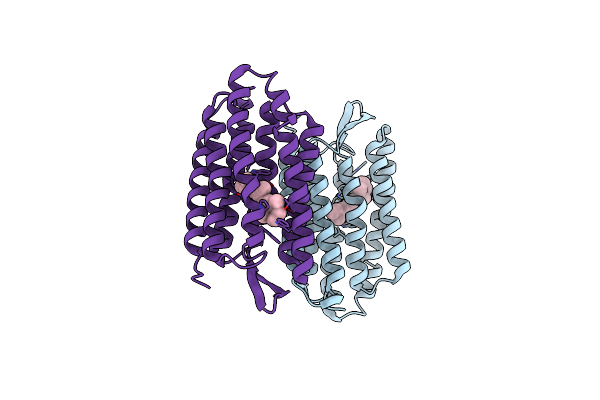
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-04-19 Classification: MEMBRANE PROTEIN Ligands: RET, D12, D10, C14, OCT, CPS |
 |
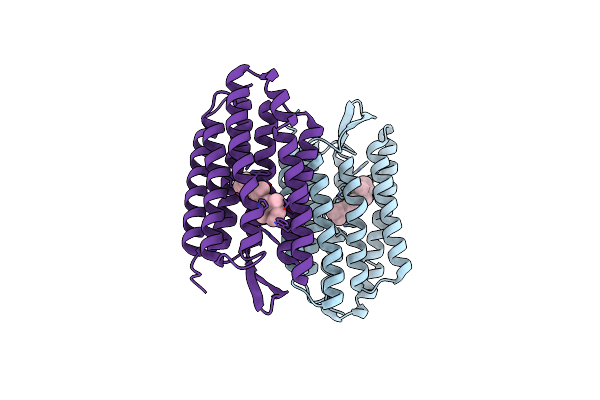
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-10-19 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
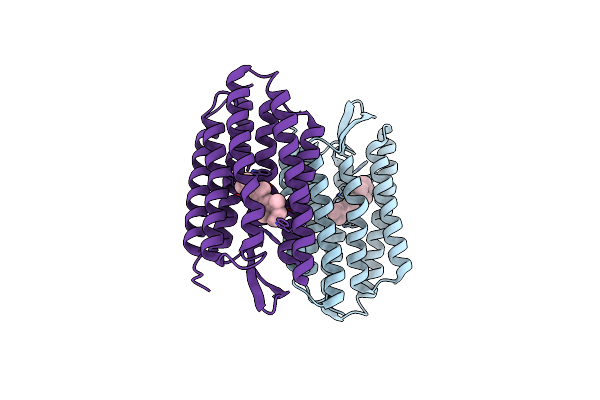
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-10-12 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-03-02 Classification: PROTON TRANSPORT Ligands: RET |
 |
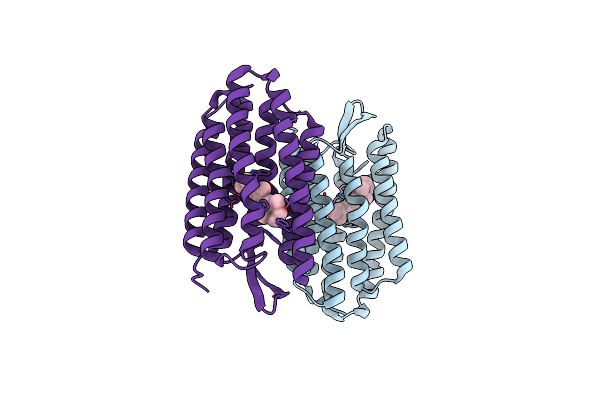
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-03-02 Classification: PROTON TRANSPORT Ligands: RET |
 |
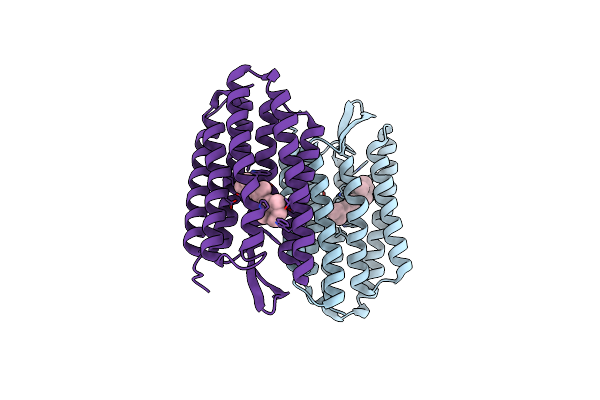
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-03-02 Classification: PROTON TRANSPORT Ligands: RET |
 |
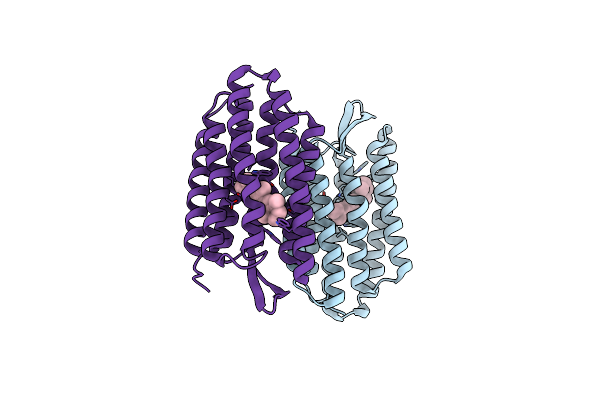
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-02 Classification: PROTON TRANSPORT Ligands: RET |
 |
Crystal Structure Of Bacteriorhodopsin Mutant P186A Crystallized From Bicelles
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-01-06 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
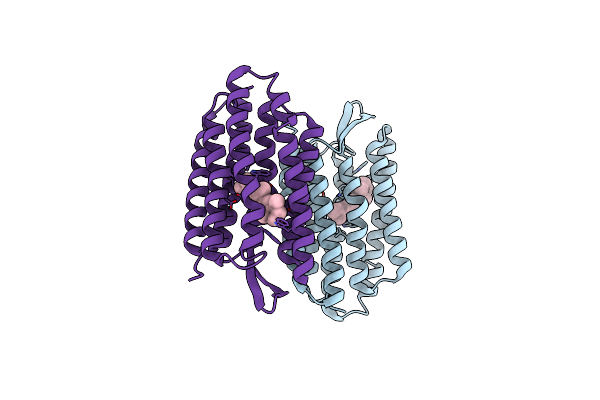
Crystal Structure Of Bacteriorhodopsin Mutant P91A Crystallized From Bicelles
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-01-06 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
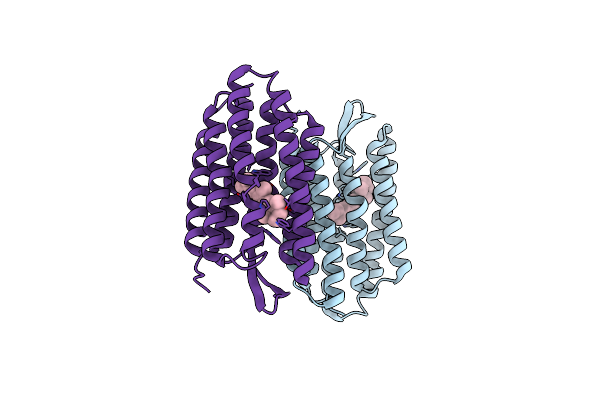
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-12-16 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-12-16 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-12-16 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Crystal Structures Of The N-Terminal Fragment From Moloney Murine Leukemia Virus Reverse Transcriptase Complexed With Nucleic Acid: Functional Implications For Template-Primer Binding To The Fingers Domain
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2000-04-04 Classification: TRANSFERASE/DNA |
 |
Use Of An N-Terminal Fragment From Moloney Murine Leukemia Virus Reverse Transcriptase To Facilitate Crystallization And Analysis Of A Pseudo-16-Mer Dna Molecule Containing G-A Mispairs
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-04-02 Classification: HYDROLASE/DNA |
 |
Crystal Structures Of The N-Terminal Fragment From Moloney Murine Leukemia Virus Reverse Transcriptase Complexed With Nucleic Acid: Functional Implications For Template-Primer Binding To The Fingers Domain
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-04-02 Classification: TRANSFERASE/DNA |
 |
Crystal Structures Of The N-Terminal Fragment From Moloney Murine Leukemia Virus Reverse Transcriptase Complexed With Nucleic Acid: Functional Implications For Template-Primer Binding To The Fingers Domain
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-03-20 Classification: TRANSFERASE/DNA Ligands: HG |

