Search Count: 15
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: ZN, AV0 |
 |
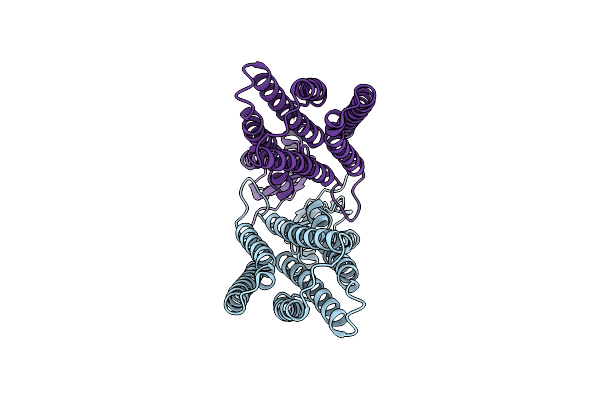
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: REPLICATION |
 |
Structure Of Mcm2-7 Dh Complexed With Cdc7-Dbf4 In The Presence Of Atpgs, State Iii
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: REPLICATION Ligands: AGS, MG, ZN, ADP |
 |
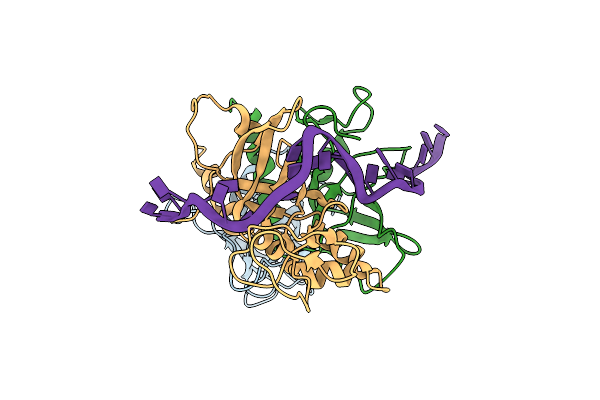
Structure Of Mcm2-7 Dh Complexed With Cdc7-Dbf4 In The Presence Of Adp:Bef3, State I
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: REPLICATION Ligands: ADP, MG, ZN, BEF |
 |
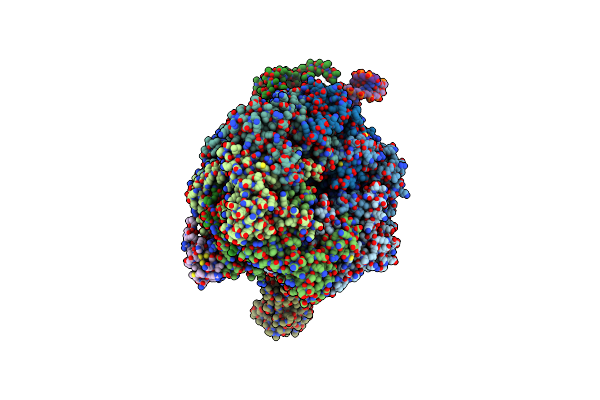
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: DNA BINDING PROTEIN |
 |
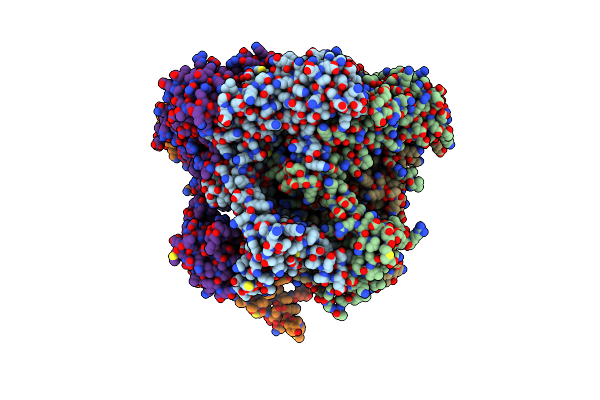
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN Ligands: ADP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-12-13 Classification: GENE REGULATION Ligands: ADP |
 |
Crystal Structure Of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant)
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-04 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Vitamin D Hydroxylase Cytochrome P450 105A1 (R73A/R84A Mutant) In Complex With 1Alpha,25-Dihydroxyvitamin D3
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-11-04 Classification: OXIDOREDUCTASE Ligands: HEM, VDX |
 |
Crystal Structure Of Vitamin D Hydroxylase Cytochrome P450 105A1 (Wild Type) With Imidazole Bound
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-04-08 Classification: OXIDOREDUCTASE Ligands: HEM, IMD |
 |
Crystal Structure Of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84A Mutant)
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-04-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84A Mutant) In Complex With 1,25-Dihydroxyvitamin D3
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-08 Classification: OXIDOREDUCTASE Ligands: HEM, VDX |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2007-10-30 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: C2E |

