Search Count: 29
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-08-31 Classification: UNKNOWN FUNCTION Ligands: BR |
 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-08-14 Classification: HYDROLASE Ligands: NAG, EDO, K |
 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-08-14 Classification: HYDROLASE Ligands: NAG, X6X |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2017-10-04 Classification: STRUCTURAL PROTEIN |
 |
Structure Of The Alg44 Pilz Domain From Pseudomonas Aeruginosa Pao1 In Complex With C-Di-Gmp
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-04-08 Classification: PROTEIN BINDING Ligands: C2E, EDO |
 |
Structure Of The Alg44 Pilz Domain (R95A Mutant) From Pseudomonas Aeruginosa Pao1 In Complex With C-Di-Gmp
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-08 Classification: PROTEIN BINDING Ligands: C2E, CL, NA |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-08-25 Classification: PROTEIN BINDING Ligands: CL, GOL |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
Penicillium Citrinum Alpha-1,2-Mannosidase In Complex With A Substrate Analog
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Escherichia Coli Adenylosuccinate Lyase Mutant H171N With Bound Amp And Fumarate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-07-03 Classification: LYASE Ligands: FUM, AMP |
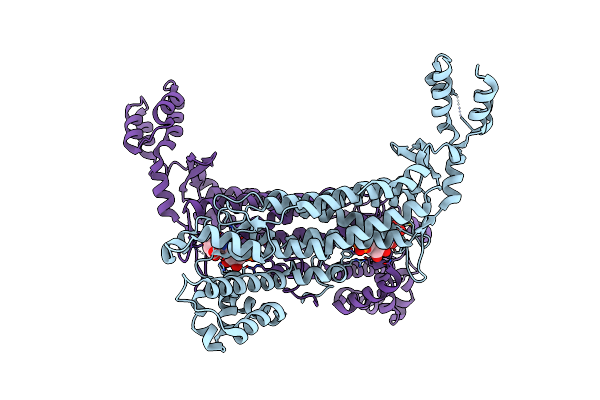 |
Crystal Structure Of Escherichia Coli Adenylosuccinate Lyase Mutant H171A With Bound Adenylosuccinate Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-07-03 Classification: LYASE Ligands: 2SA |
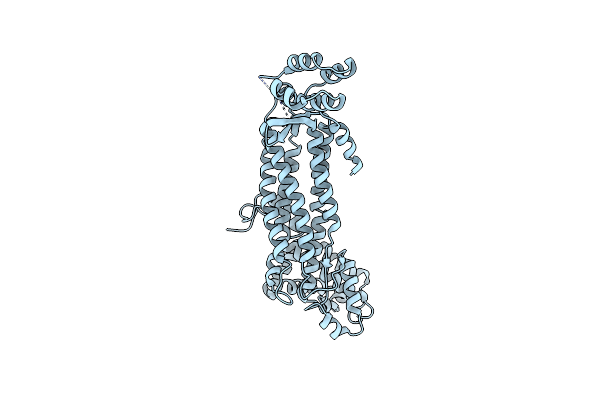 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-07-03 Classification: LYASE |
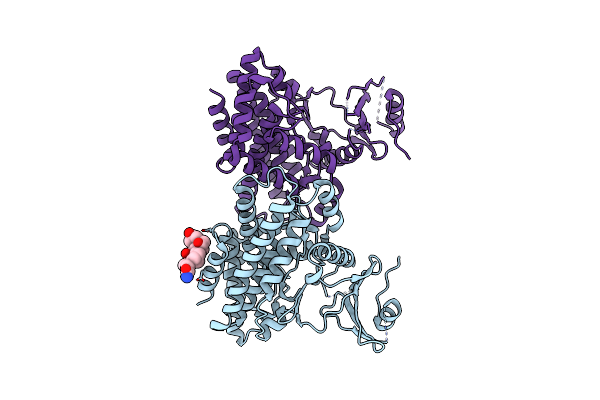 |
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: CPS |
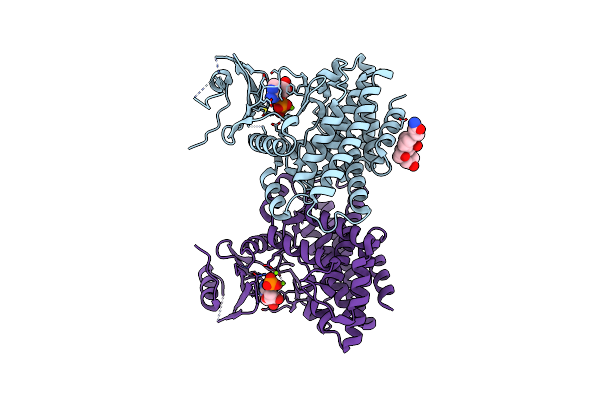 |
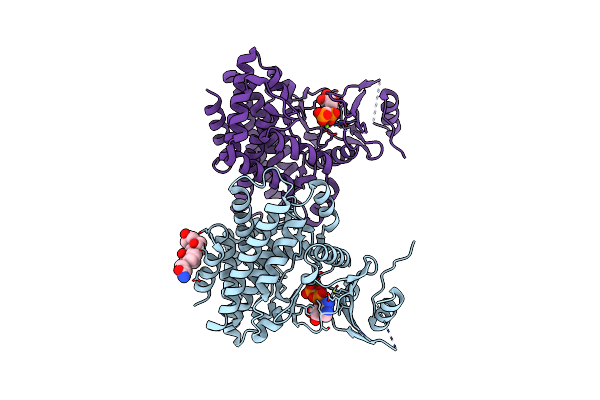
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, ADP, CPS |
 |
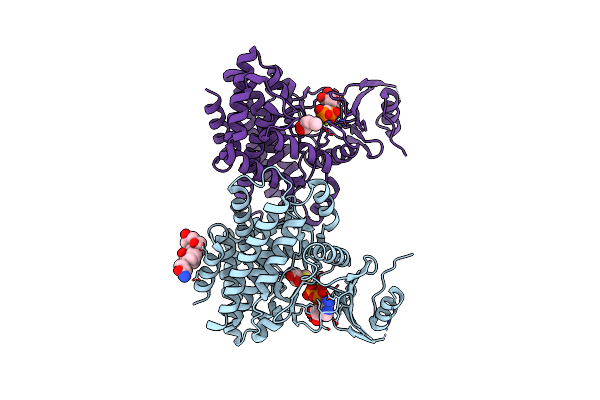
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, CPS, ACP |
 |
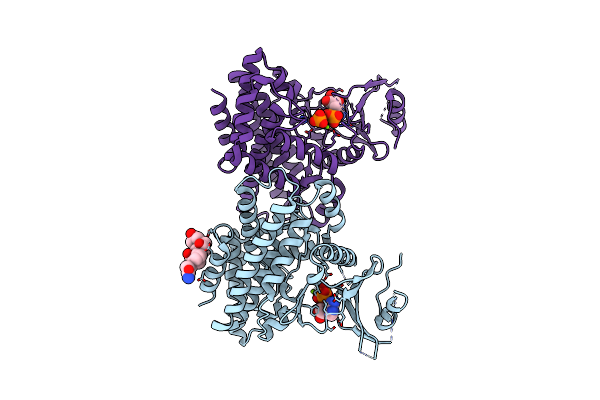
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, CPS, ACP, SR1 |
 |
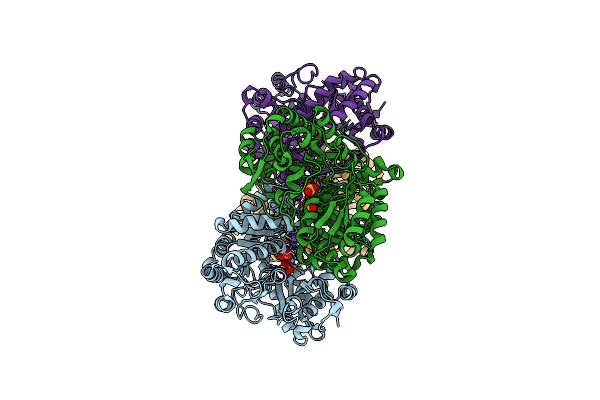
Structures Of 5-Methylthioribose Kinase Reveal Substrate Specificity And Unusual Mode Of Nucleotide Binding
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: MG, ADP, PO4, CPS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-06-28 Classification: LYASE Ligands: SO4, K |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-05-04 Classification: TRANSFERASE Ligands: CL, GOL |
 |
Crystal Structure Of Yeast Alpha1,2-Mannosyltransferase Kre2P/Mnt1P: Binary Complex With Gdp/Mn
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-05-04 Classification: TRANSFERASE Ligands: MN, CL, GDP, EPE |

