Search Count: 126
 |
Organism: Citrus sinensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PLANT PROTEIN Ligands: D12 |
 |
Organism: Citrus sinensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PLANT PROTEIN Ligands: D12, R16 |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Clostridium sp. of09-36
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: GENE REGULATION Ligands: O6E |
 |
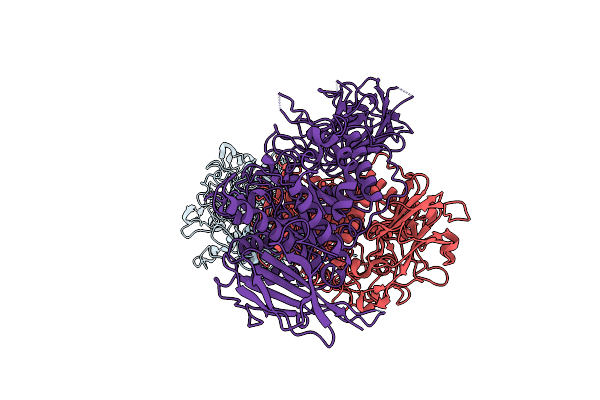
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: HYDROLASE |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: HYDROLASE |
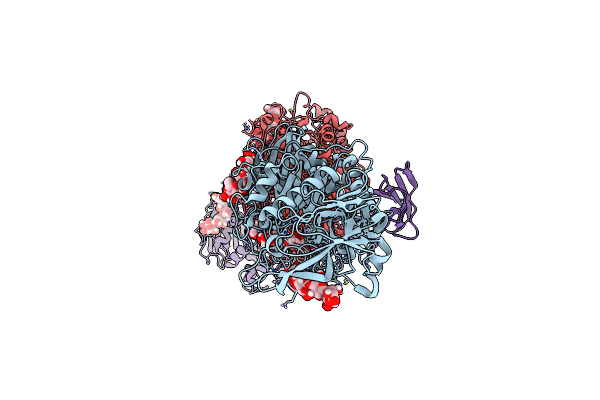 |
Cryo-Em Structure Of The Rice Isoamylase Isa1-Isa2 Heterocomplex Incubated With Amylopectin
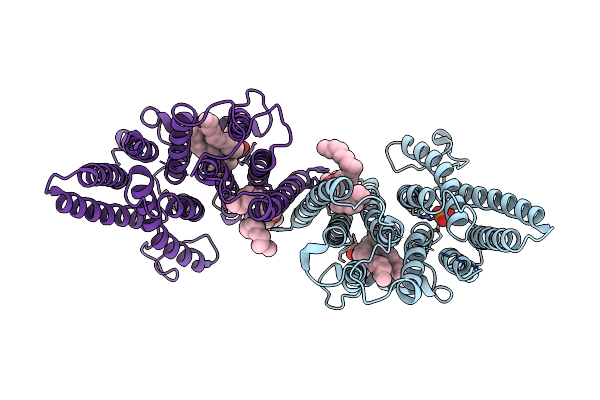
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GLC |
 |
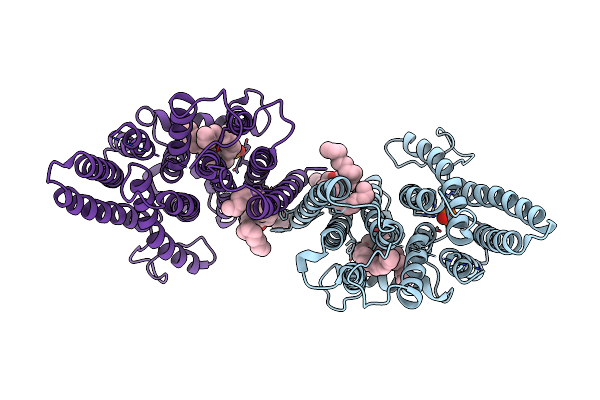
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSPORT PROTEIN Ligands: PC1, PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSPORT PROTEIN Ligands: PC1, PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSPORT PROTEIN Ligands: PC1, PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: TRANSPORT PROTEIN Ligands: PC1 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-09-04 Classification: LIPID BINDING PROTEIN Ligands: H9X |
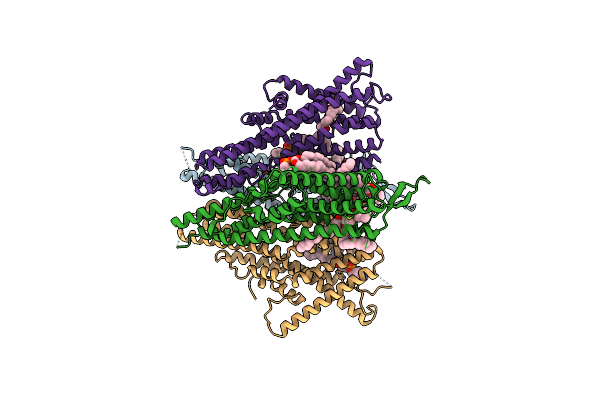 |
Cryo-Em Structure Of The Insect Olfactory Receptor Or5-Orco Heterocomplex From Acyrthosiphon Pisum Bound With Geranyl Acetate
Organism: Acyrthosiphon pisum
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: PC1, SOU |
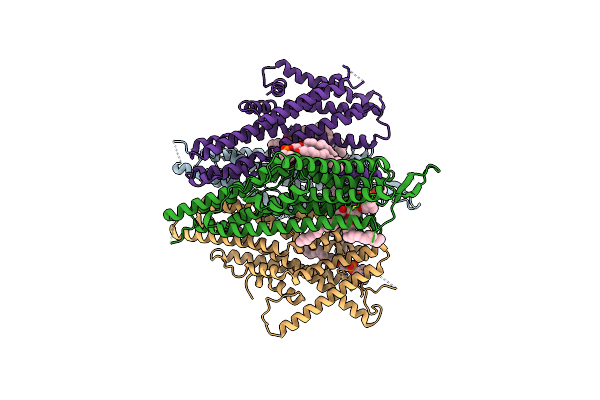 |
Cryo-Em Structure Of The Insect Olfactory Receptor Or5-Orco Heterocomplex From Acyrthosiphon Pisum
Organism: Acyrthosiphon pisum
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: PC1 |
 |
Cryo-Em Structure Of The Yeast Tom Core Complex Crosslinked By Bs3 (From Tom-Tim23 Complex)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: TRANSLOCASE Ligands: 46E |

