Search Count: 81
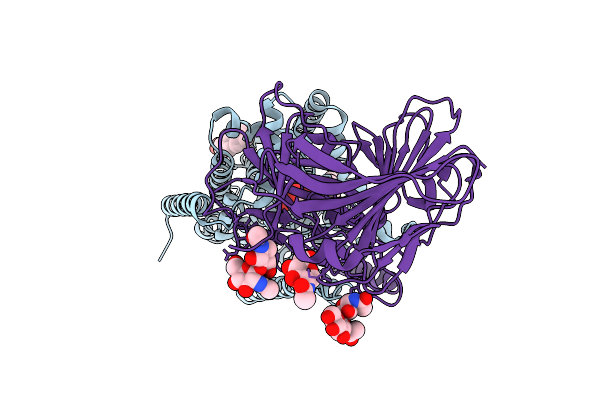 |
Cryo-Em Structure Of Candida Glabrata Gpi Mannosyltransferase I Bound To Dol-P-Man
Organism: Nakaseomyces glabratus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: MJC, Y01, NAG |
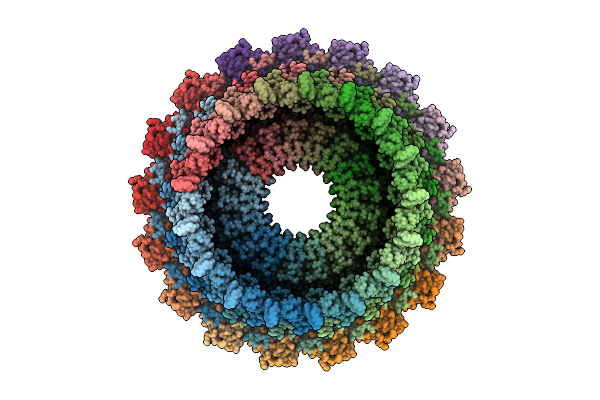 |
Organism: Escherichia coli o127:h6 str. e2348/69
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
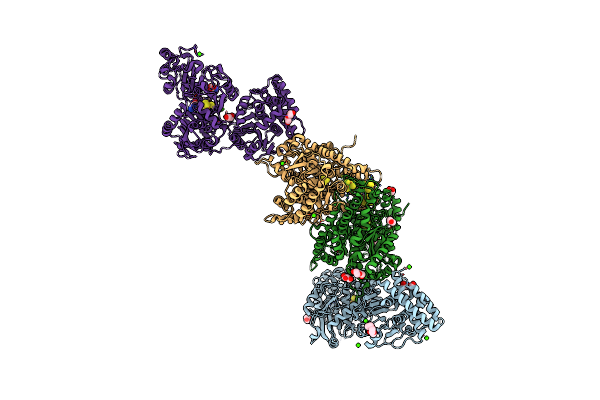 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Acetyl-Coenyzme A From Clostridium Autoethanogenum
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: SF4, NI, EDO, PE4, GOL, PEG, CA, CL, XCC, ACO, TRS |
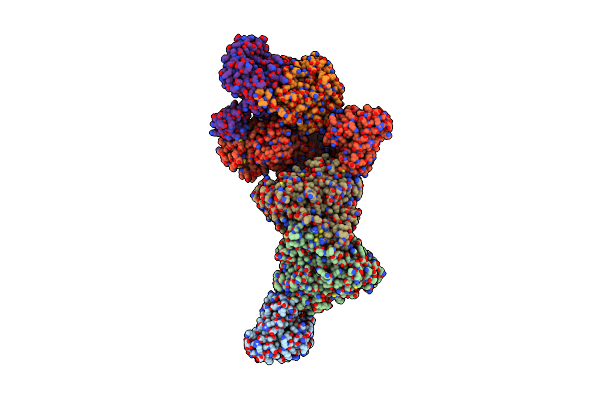 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3A)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, B12, NI, RQM |
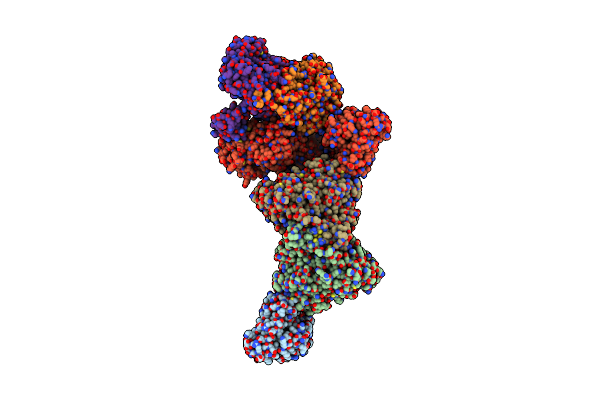 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3B)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM, B12 |
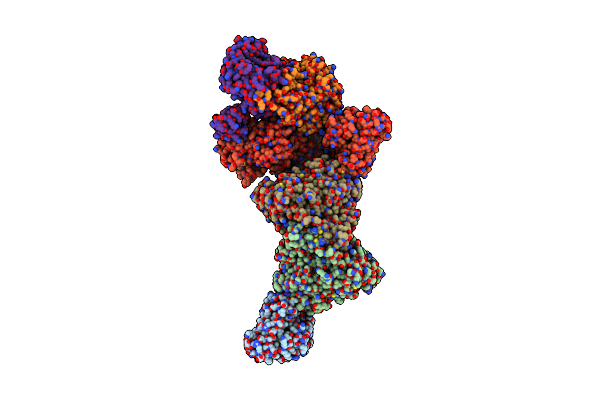 |
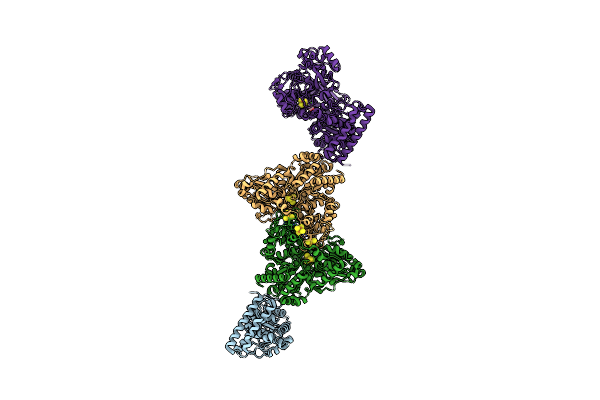
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Corrinoid Iron-Sulfur Protein (Cofesp) From Clostridium Autoethanogenum (Composite Structure, Class 3Cb)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, B12, RQM |
 |
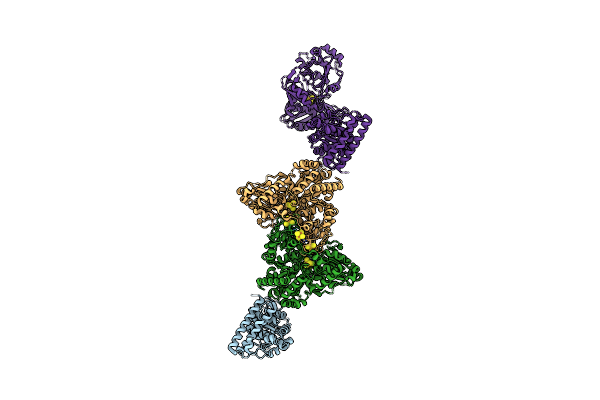
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Closed And Co-Bound State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM, NI, CMO |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum (Composite Structure, Semi-Extended State)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, NI, RQM |
 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Ferredoxin (Clostridium Autoethanogenum)
Organism: Clostridium autoethanogenum dsm 10061
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: SF4, RQM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: A1L26 |
 |
Organism: Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: A1L26, TPF |
 |
Organism: Candida albicans sc5314
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: A1L26, A1L27 |
 |
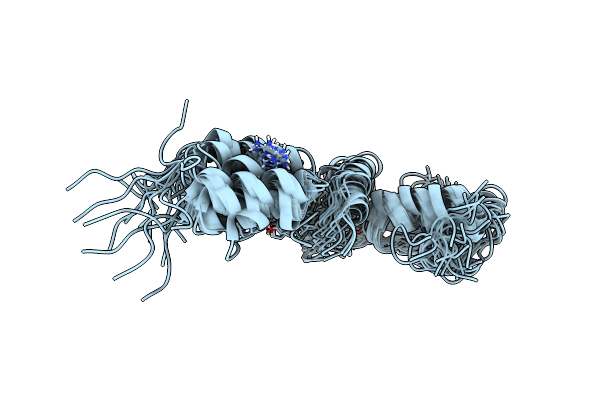
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-08-14 Classification: TRANSCRIPTION Ligands: ZN |
 |
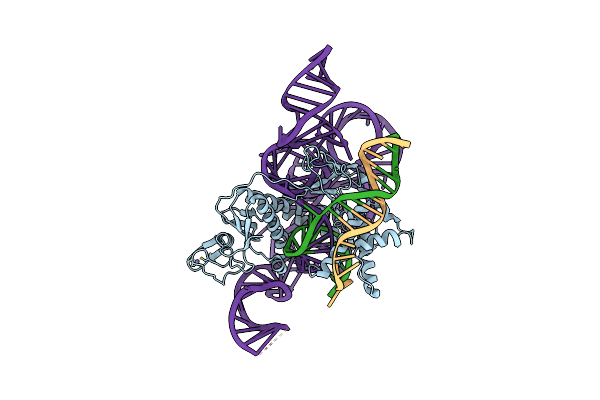
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-07-24 Classification: TRANSCRIPTION Ligands: ZN |
 |
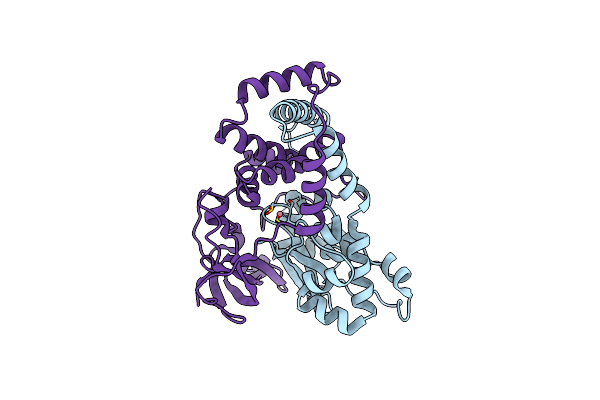
Organism: Firmicutes bacterium am43-11bh
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Mutant From Pseudomonas Thermophila - L6T
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE Ligands: CO |
 |
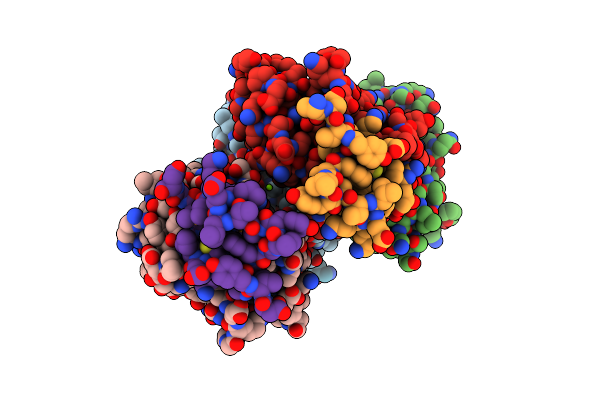
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-20 Classification: PROTEIN TRANSPORT Ligands: 1PE, GOL, CL, ACY |
 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-02-01 Classification: DNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-02-01 Classification: DNA BINDING PROTEIN Ligands: ZN |

