Search Count: 186
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: SO4, NA |
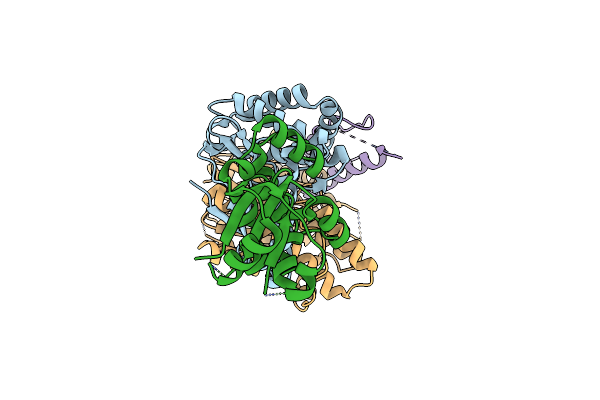 |
Organism: Methanocaldococcus jannaschii, Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: RNA BINDING PROTEIN |
 |
Structure Of The Keops Complex (Cgi121/Bud32/Kae1/Pcc1) Bound To Trna In Its Native-Like Conformation.
Organism: Methanocaldococcus jannaschii, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: RNA BINDING PROTEIN/RNA |
 |
Structure Of The Keops Complex (Cgi121/Bud32/Kae1/Pcc1) Bound To Trna In A Distorted Trna Conformation
Organism: Methanocaldococcus jannaschii, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Human betaherpesvirus 5
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human betaherpesvirus 5
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-10 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The Methyltransferase Domain Of R882H/R676K Dnmt3A Homotetramer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: SAH, GOL, TLA |
 |
Crystal Structure Of The Methyltransferase Domain Of R882C/R676K Dnmt3A Homotetramer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The Methyltransferase Domain Of R882H/N879A Dnmt3A Homotetramer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: MEMBRANE PROTEIN Ligands: MG, HP6, C14 |
 |
Crystal Structure Of The Mixed Lineage Leukaemia (Mll1) Set Domain With The Cofactor Product S-Adenosylhomocysteine And Borealin Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-09-27 Classification: GENE REGULATION Ligands: ZN, SAH |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: PROTEIN TRANSPORT Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: PROTEIN TRANSPORT Ligands: NAG, CLR |

