Search Count: 129
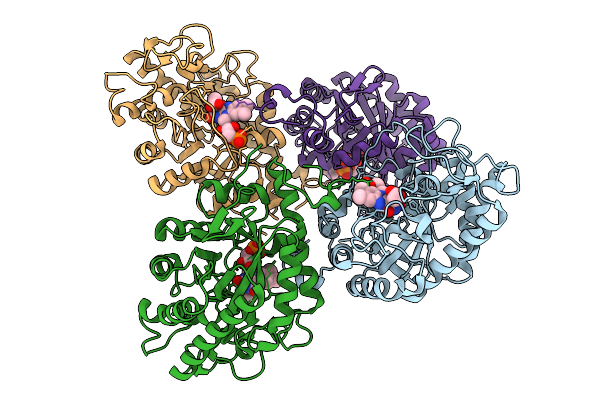 |
Organism: Parageobacillus thermantarcticus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: ACT, FMN |
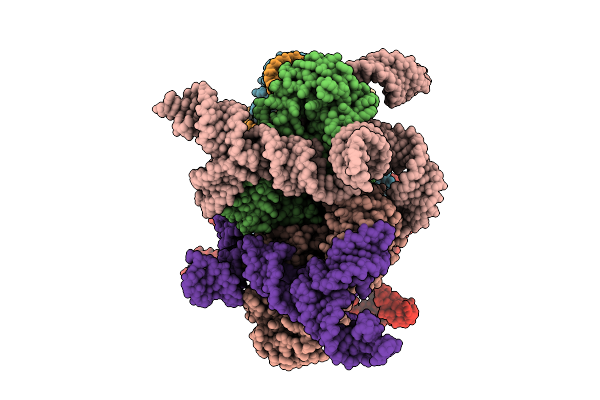 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: ANTIVIRAL PROTEIN/RNA Ligands: MG |
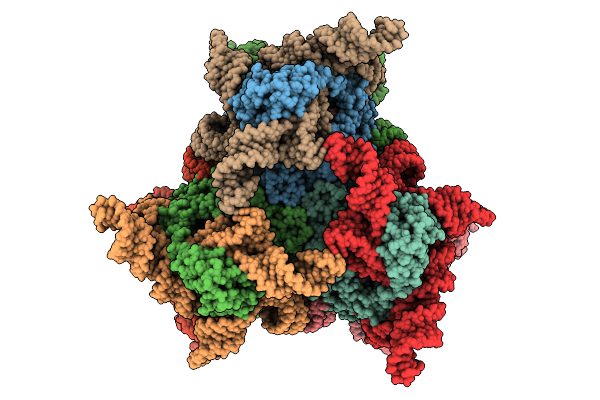 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: ANTIVIRAL PROTEIN/RNA/DNA Ligands: DTP, MG |
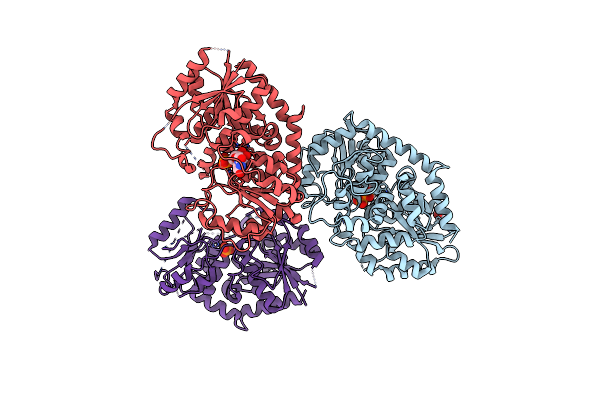 |
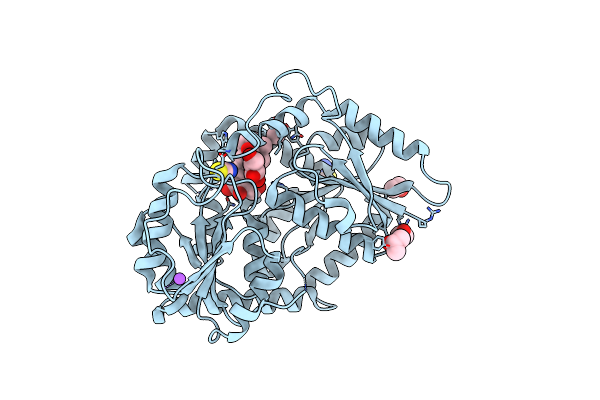
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSFERASE Ligands: UPG, IPA, PO4 |
 |
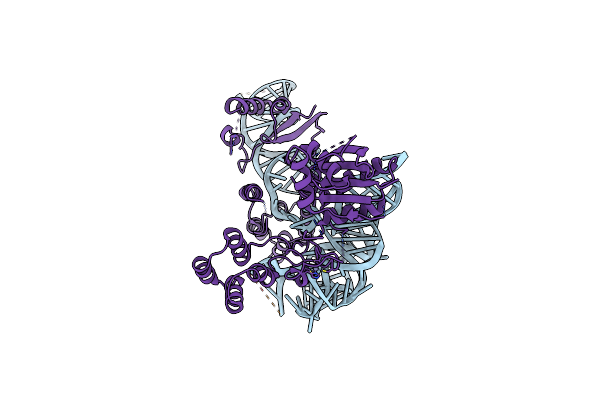
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
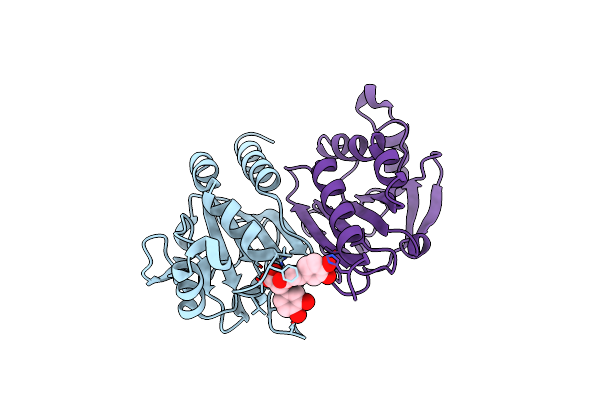
Organism: Metagenome
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
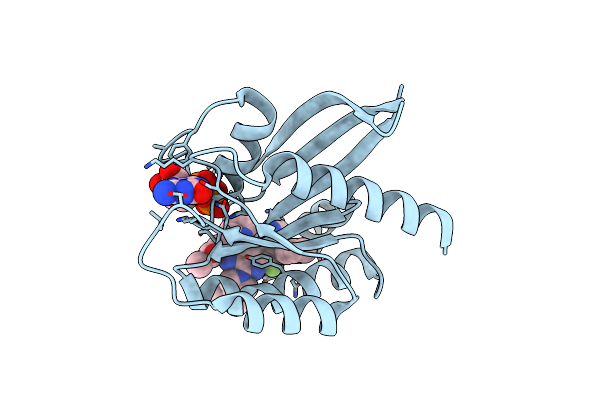
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-22 Classification: OXIDOREDUCTASE Ligands: ROA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, MG, A1BEI |
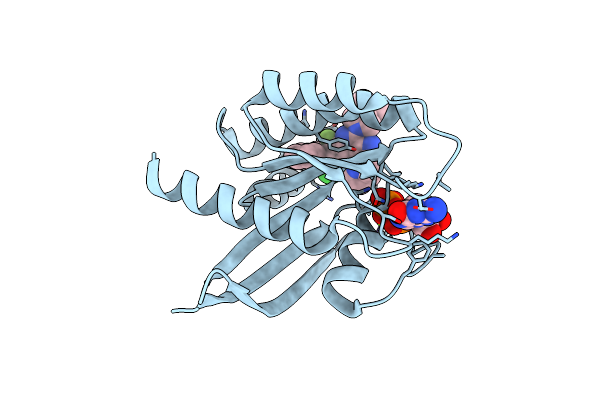 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1BEJ, MG |
 |
Organism: Homo sapiens, Homo heidelbergensis, Severe acute respiratory syndrome coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
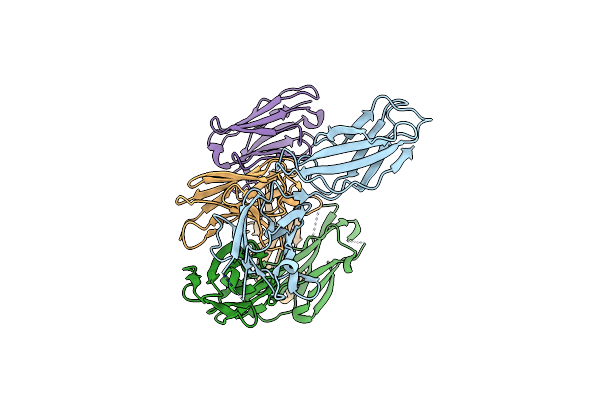 |
Structure Of Interleukin Receptor Common Gamma Chain (Il2Rgamma/Cd132) In Complex With 2D4
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN |
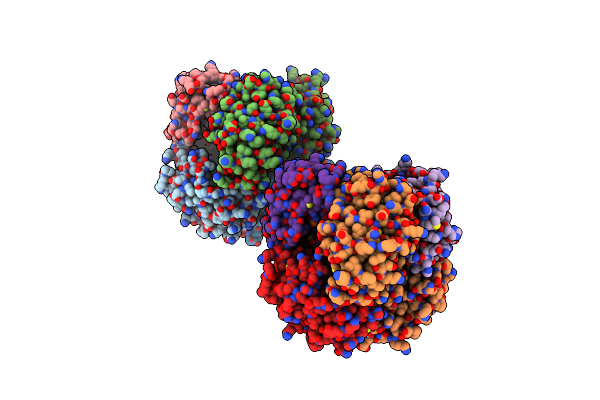 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: ATP |
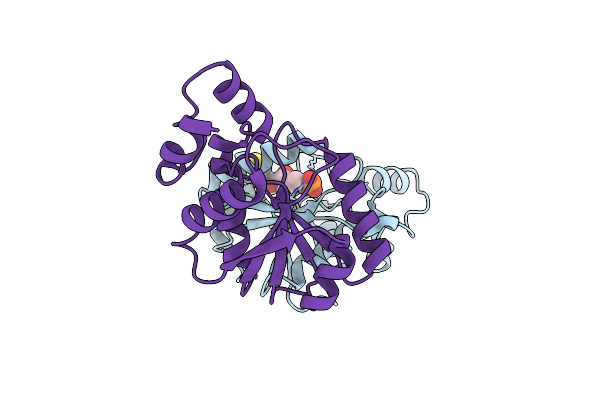 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-30 Classification: TRANSFERASE |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: COD |
 |
Organism: Selenomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: ANTIVIRAL PROTEIN/DNA/RNA |
 |
Organism: Selenomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: ANTIVIRAL PROTEIN/DNA/RNA |
 |
Organism: Selenomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Selenomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: ANTIVIRAL PROTEIN |
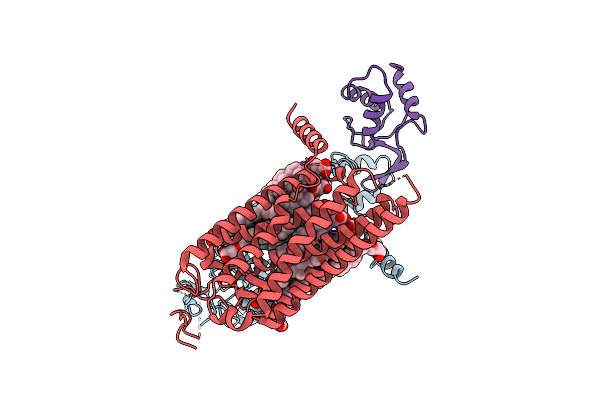 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: Y01 |

