Search Count: 41
 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: ILE |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Mutant From Pseudomonas Thermophila - L6T
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE Ligands: CO |
 |
Organism: Exiguobacterium sibiricum (strain dsm 17290 / cip 109462 / jcm 13490 / 255-15)
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: GOL, CA |
 |
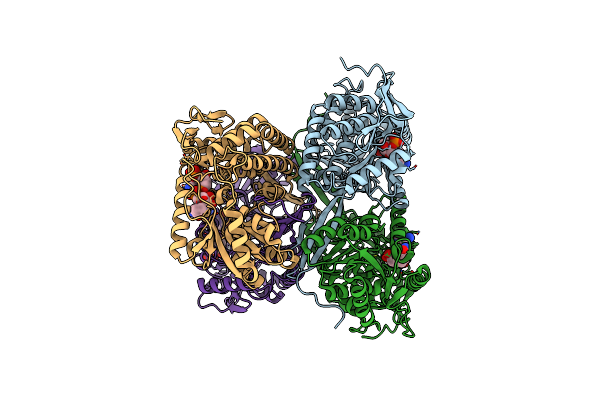
The C296A Mutant Of L-Sorbosone Dehydrogenase (Sndh) From Gluconobacter Oxydans Wsh-004
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-03-01 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
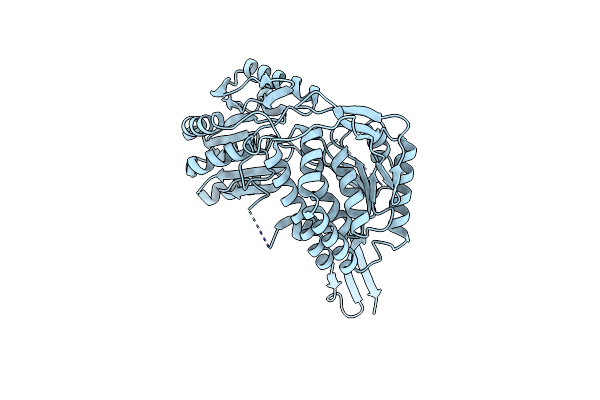
The Crystal Structure Of The Reduced Form Of Gluconobacter Oxydans Wsh-004 Sndh
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2023-03-01 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
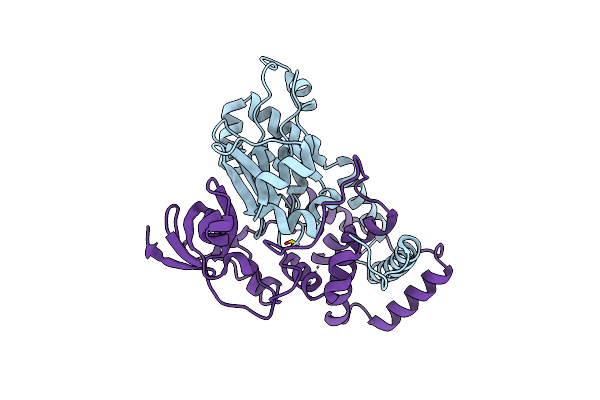
The Crystal Structure Of The Oxidized Form Of Gluconobacter Oxydans Wsh-004 Sndh
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: OXIDOREDUCTASE |
 |
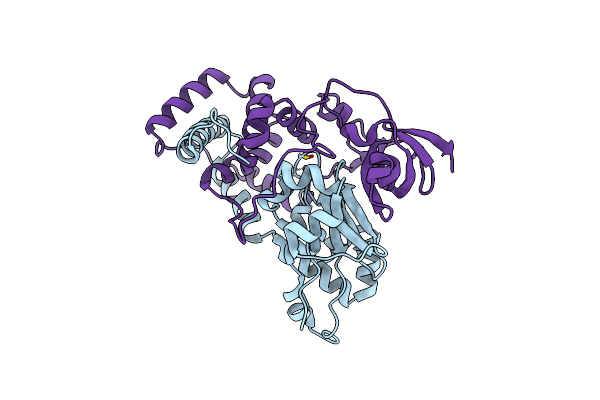
Crystal Structure Of Co-Type Nitrile Hydratase Mutant From Pseudonocardia Thermophila - M46R
Organism: Pseudonocardia thermophila dsm 43832
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-08-24 Classification: LYASE Ligands: CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Mutant From Pseudomonas Thermophila - A129R
Organism: Pseudonocardia thermophila dsm 43832, Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-24 Classification: LYASE Ligands: CO |
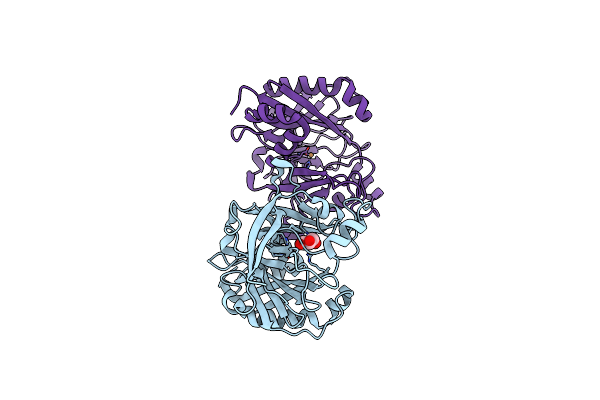 |
Crystal Structures Of 2-Oxoglutarate Dependent Dioxygenase (Ctb9) From Cercospora Sp. Jnu001
Organism: Cercospora sojina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
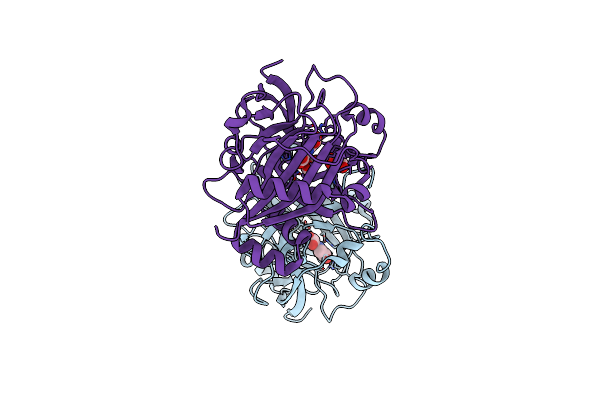 |
Crystal Structures Of 2-Oxoglutarate Dependent Dioxygenase (Ctb9) In Complex With N-Oxalylglycine
Organism: Cercospora sojina
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE Ligands: CU, OGA, EDO, GOL |
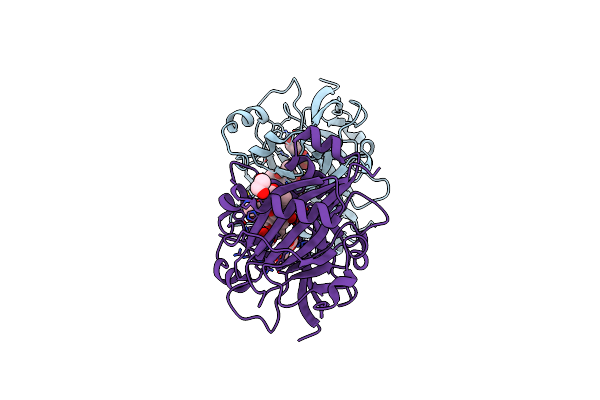 |
Crystal Structures Of 2-Oxoglutarate Dependent Dioxygenase (Ctb9) In Complex With N-Oxalylglycine And Pre-Cercosporin
Organism: Cercospora sojina
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE Ligands: CU, OGA, JD9, EDO |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Inhibited State Sb_Usp14
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: HYDROLASE Ligands: ATP, ADP, ZN |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Inhibited State Sd5_Usp14
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: HYDROLASE Ligands: ATP, ADP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, MG, ADP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, MG, ADP, ZN |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, MG, ADP, ZN |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Engaged State Ed4_Usp14
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ADP, ATP, ZN |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Engaged State Ed5_Usp14
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, ADP, ZN |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Engaged State Ed0_Usp14
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, ADP, ZN |
 |
Structure Of Usp14-Bound Human 26S Proteasome In Substrate-Engaged State Ed1_Usp14
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ATP, ADP, ZN |

