Search Count: 15
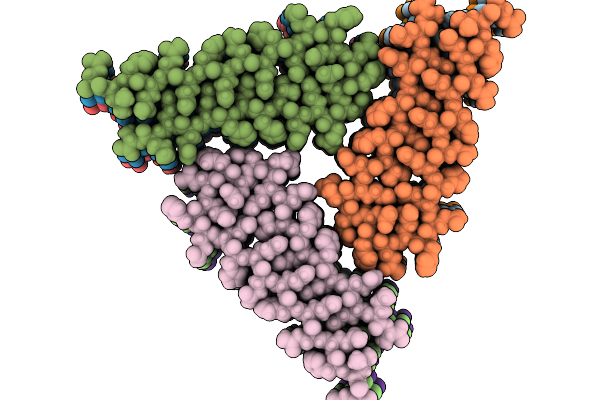 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: PROTEIN FIBRIL |
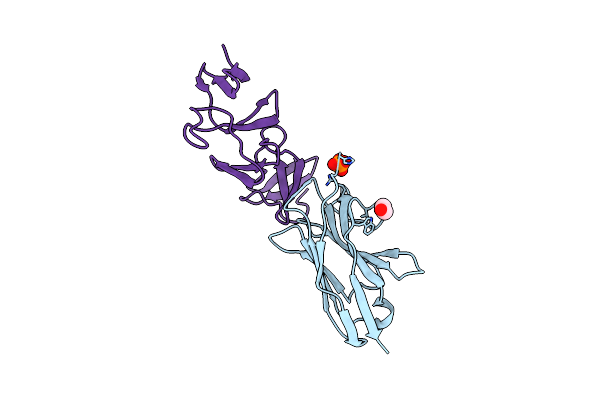 |
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2592-2710 Bound To H5.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile 342
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO, PO4 |
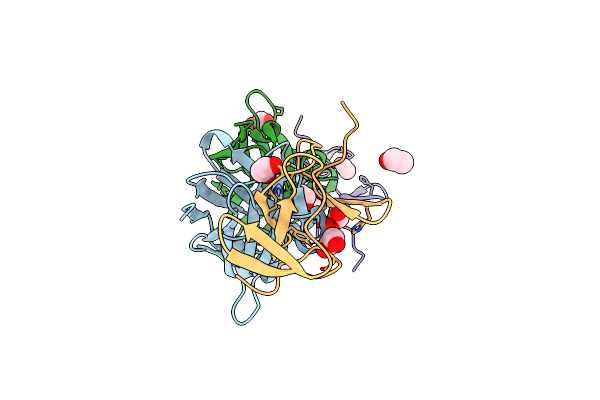 |
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2639-2707 Bound To C4.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO |
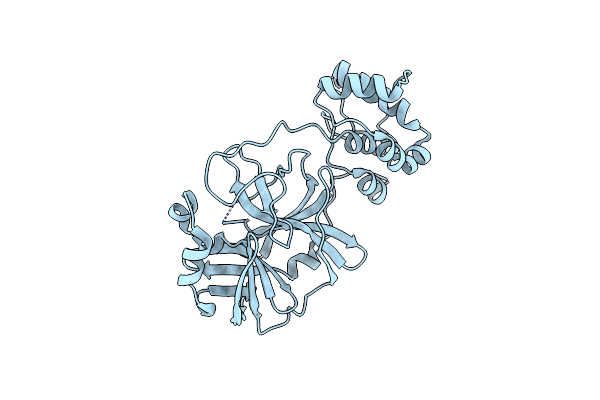 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-06-29 Classification: VIRAL PROTEIN |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: IOD |
 |
The Solution Structure Of The Micelle-Associated Fatc Domain Of The Human Protein Kinase Ataxia Telangiectasia Mutated (Atm)
Organism: Streptococcus sp. group g, Homo sapiens
Method: SOLUTION NMR Release Date: 2019-03-27 Classification: TRANSFERASE |
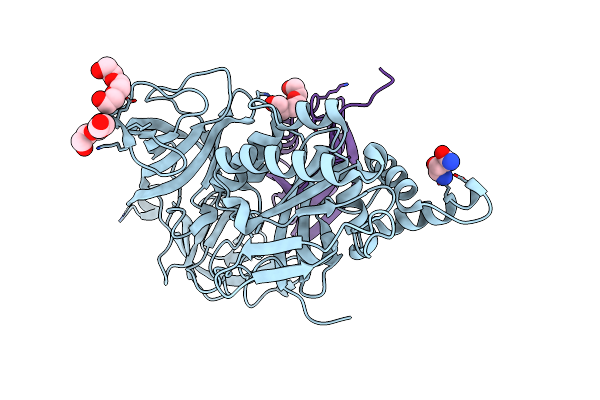 |
Crystal Structure Analysis Of The Yeast Elongation Factor Complex Eef1A:Eef1Ba
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-08-23 Classification: TRANSLATION Ligands: GLN, PGE |
 |
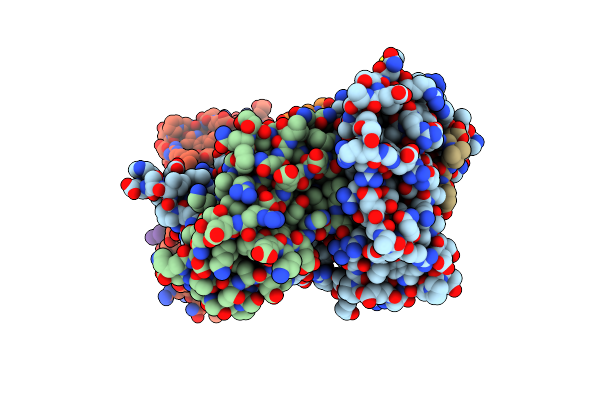
Organism: Streptomyces leeuwenhoekii
Method: SOLUTION NMR Release Date: 2015-10-07 Classification: CELL INVASION |
 |
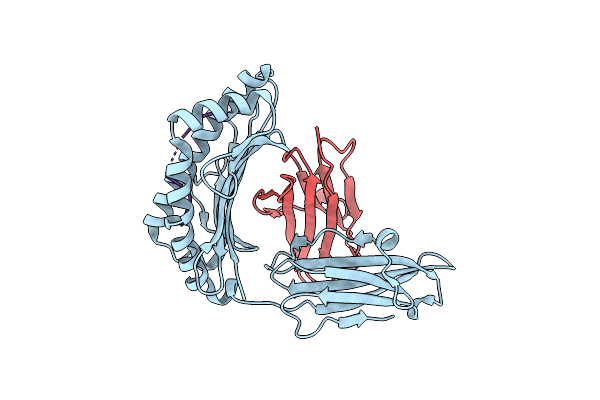
Crystal Structure Of Hla-A*2402 Complexed With A Newly Identified Peptide From 2009 H1N1 Pa (649-658)
Organism: Homo sapiens, Influenza a virus h3n2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-10-10 Classification: IMMUNE SYSTEM |
 |
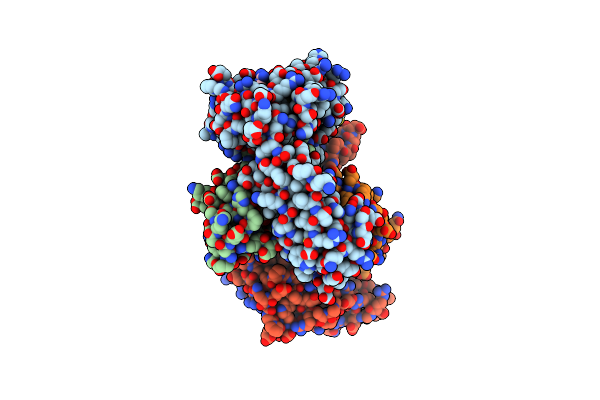
Crystal Structure Of Hla-A*2402 Complexed With A Newly Identified Peptide From 2009H1N1 Pb1 (496-505)
Organism: Homo sapiens, Influenza a virus h3n2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-10 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hla-A*2402 Complexed With A Newly Identified Peptide From 2009 H1N1 Pb1 (498-505)
Organism: Homo sapiens, Influenza a virus h3n2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: IMMUNE SYSTEM |
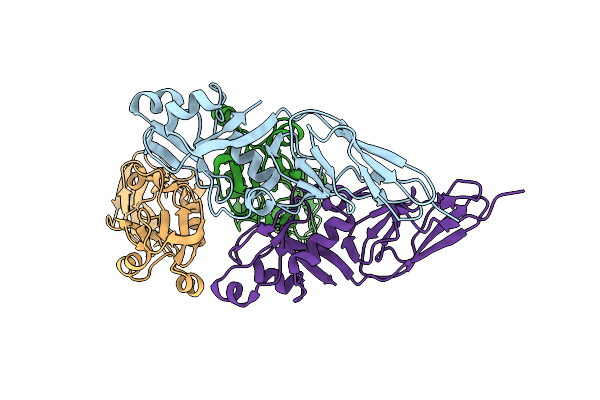 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-01-05 Classification: CELL ADHESION |
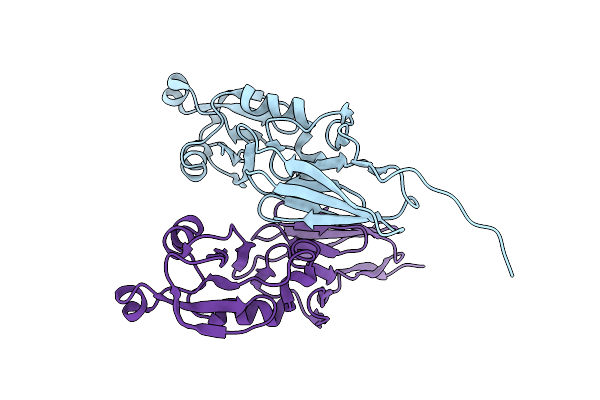 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-01-05 Classification: CELL ADHESION |
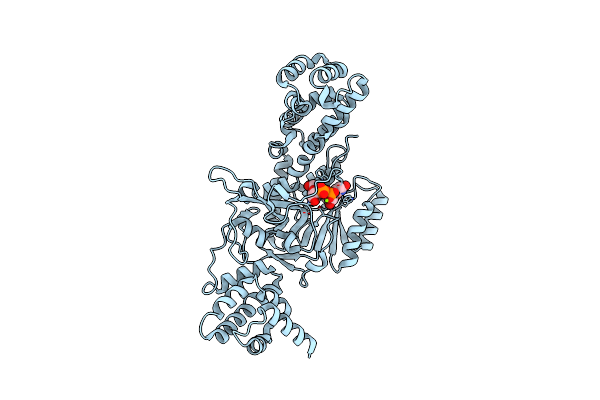 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: UPG, MG |
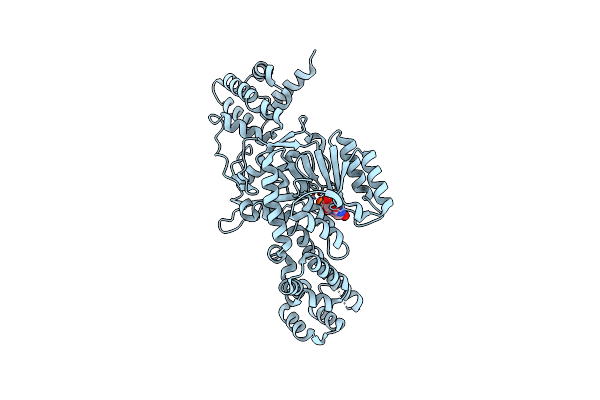 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: UDP |

