Search Count: 27
 |
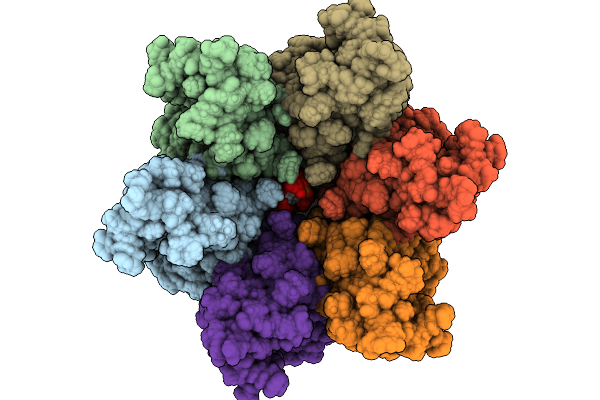
Immature Hiv-1 Cactd-Sp1 Lattice With Maturation Inhibitor Pf-46396 (R) And Inositol Hexakisphosphate (Ip6)
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: SOLID-STATE NMR Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: IHP, A1CCY |
 |
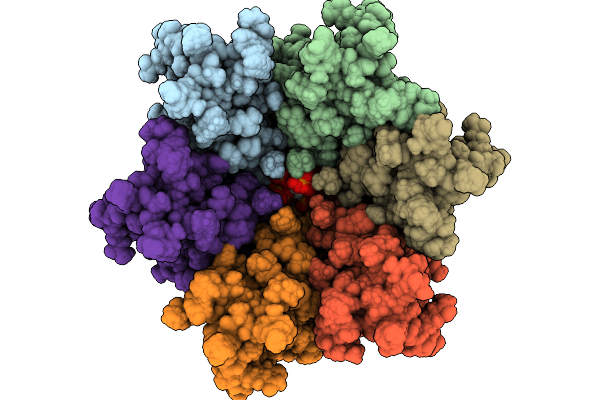
Immature Hiv-1 Cactd-Sp1 Lattice With Maturation Inhibitor Pf-46396 (R) And Inositol Hexakisphosphate (Ip6)
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: SOLID-STATE NMR Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: A1CCY, IHP |
 |
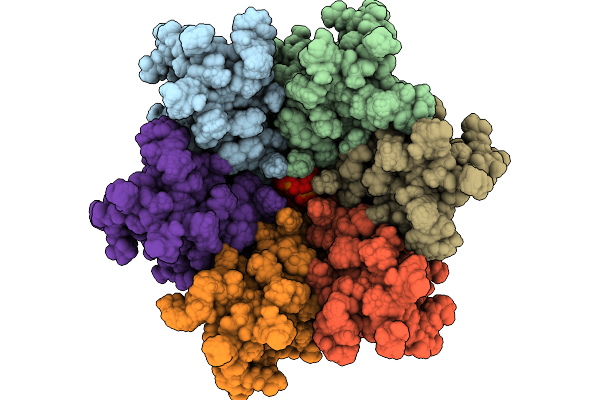
Immature Hiv-1 Cactd-Sp1 Lattice With Maturation Inhibitor Pf-46396 (S) And Inositol Hexakisphosphate (Ip6)
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: SOLID-STATE NMR Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: IHP, A1CCZ |
 |
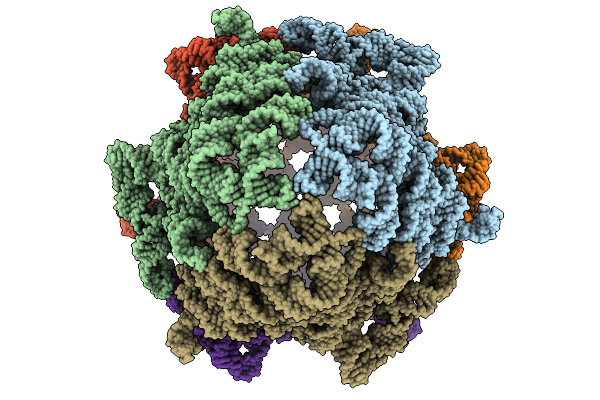
Immature Hiv-1 Cactd-Sp1 Lattice With Maturation Inhibitor Pf-46396 (S) And Inositol Hexakisphosphate (Ip6)
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: SOLID-STATE NMR Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: IHP, A1CCZ |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Uncultured caudovirales phage
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Uncultured caudovirales phage
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: RNA |
 |
Organism: Phaseolus vulgaris, Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LIGASE/RNA Ligands: ATP |
 |
Organism: Phaseolus vulgaris, Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LIGASE/RNA |
 |
Organism: Phaseolus vulgaris, Brome mosaic virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LIGASE/RNA |
 |
Crystal Structure Of 4-Fluoro-Tryptophan Labeled Oscillatoria Agardhii Agglutinin
Organism: Planktothrix agardhii
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-11-06 Classification: SUGAR BINDING PROTEIN Ligands: CXS |

