Search Count: 42
 |
The Structure And Functional Mechanism Of Nucleotide Regulated Acetylhexosaminidase Am2136 From Akkermansia Muciniphila
Organism: Akkermansia muciniphila (strain atcc baa-835 / muc)
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: MG |
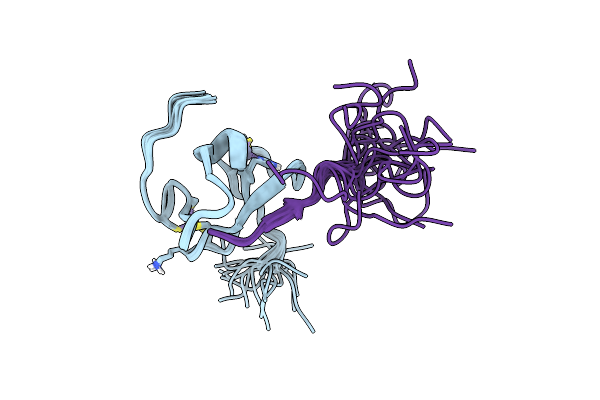 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-10-12 Classification: STRUCTURAL PROTEIN Ligands: ZN |
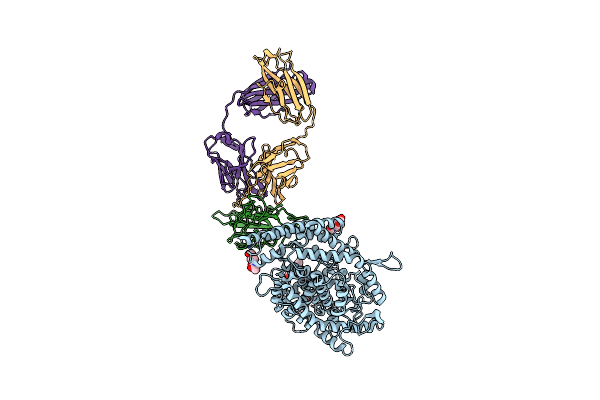 |
Sars-Cov-2 Spike (6P) In Complex With 3 R1-32 Fabs And 3 Ace2, Focused Refinement Of Rbd Region
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: ZN, NAG |
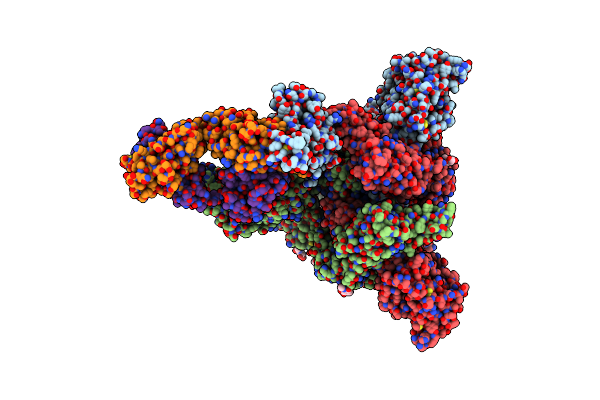 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
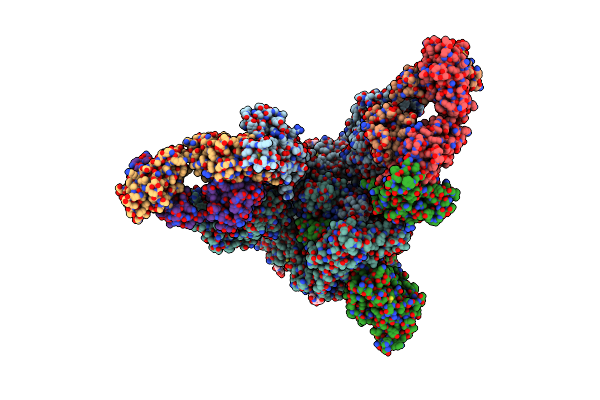 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Nanobody 17F6
Organism: Severe acute respiratory syndrome coronavirus 2, Chiloscyllium plagiosum
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-07-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain N501Y Mutant In Complex With Neutralizing Nanobody 20G6
Organism: Severe acute respiratory syndrome coronavirus 2, Chiloscyllium plagiosum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
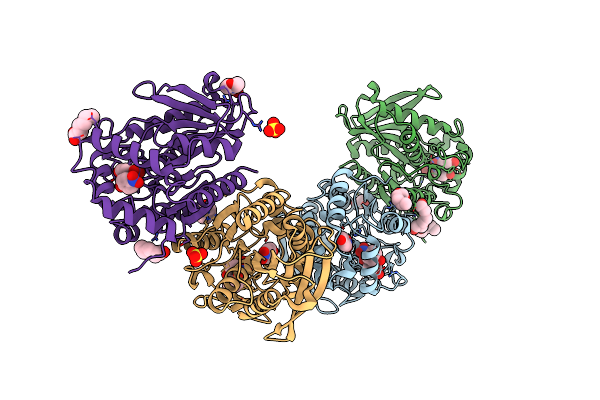
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: D8F, SO4, EDO, GOL, 6NA |
 |
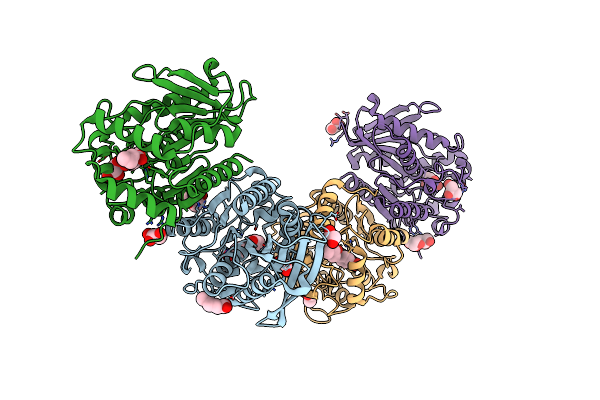
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: NPO, 6NA, PGE, EDO, SO4, MES |
 |
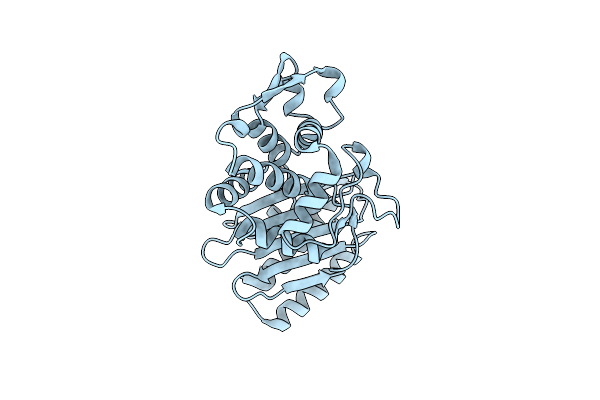
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: D8F, 6NA, EDO, GOL, 2FK |
 |
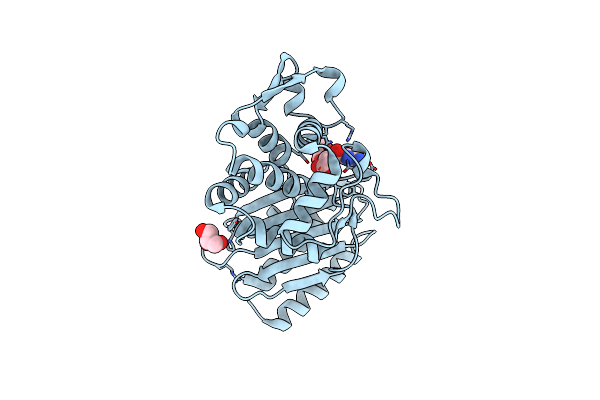
Crystal Structure Of Class A B-Lactamase, Penl, Variant Cys69Tyr, From Burkholderia Thailandensis
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-01-15 Classification: HYDROLASE |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Cys69Tyr, From Burkholderia Thailandensis, In Complex With Ceftazidime-Like Boronic Acid
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: CB4, GOL |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Asn136Asp, From Burkholderia Thailandensis
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Asn136Asp, From Burkholderia Thailandensis, In Complex With Ceftazidime-Like Boronic Acid
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: CB4, ACT |
 |
Organism: Pseudomonas aeruginosa, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-12-25 Classification: ANTIMICROBIAL PROTEIN Ligands: SO4, SPD |
 |
Organism: Chenopodium quinoa
Method: SOLUTION NMR Release Date: 2019-05-15 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-03-13 Classification: PROTEIN BINDING/SIGNALING PROTEIN |
 |
Crystal Structure Of The Gemin2-Binding Domain Of Smn, Gemin2 In Complex With Smd1(1-82)/D2/F/E/G From Human
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2018-07-04 Classification: SPLICING |

