Search Count: 16
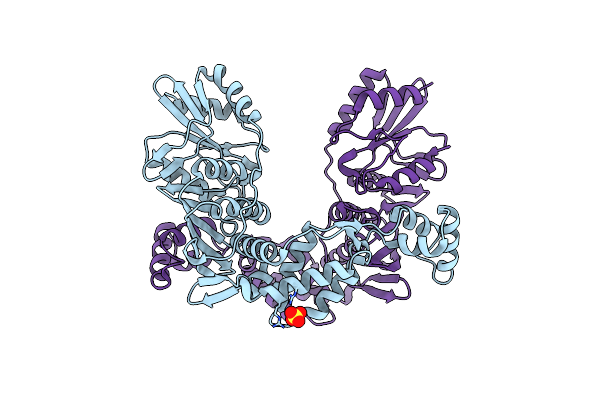 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: SO4 |
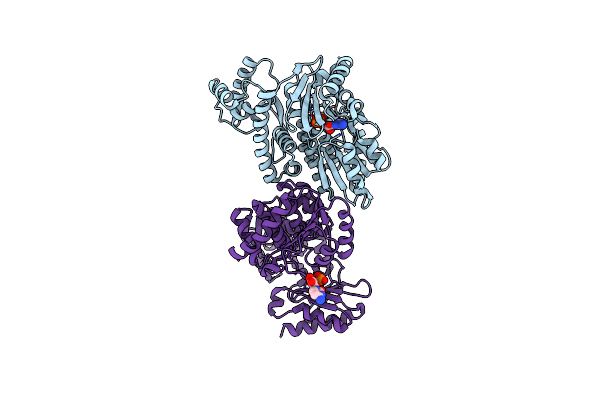 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-08-02 Classification: DNA BINDING PROTEIN Ligands: IY0 |
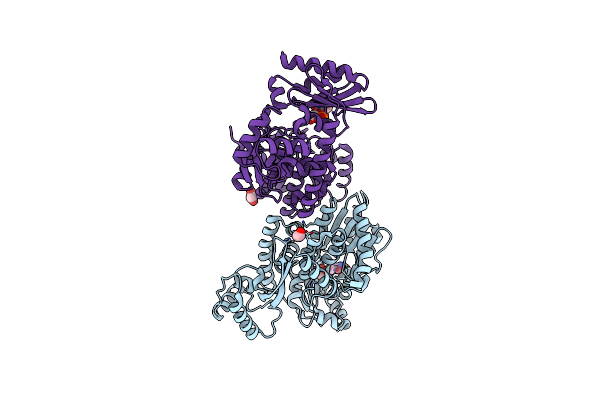 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2023-08-02 Classification: DNA BINDING PROTEIN Ligands: IY9, EDO |
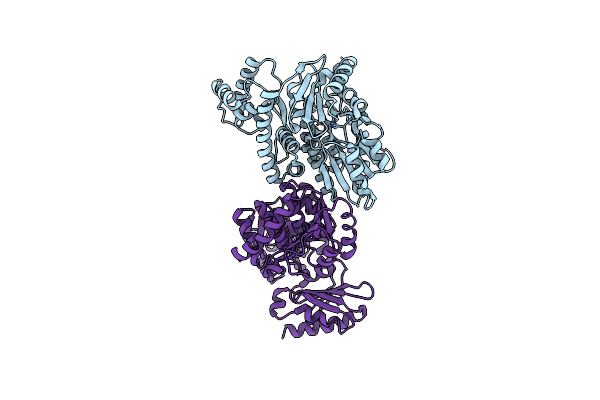 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-08-02 Classification: DNA BINDING PROTEIN Ligands: MN |
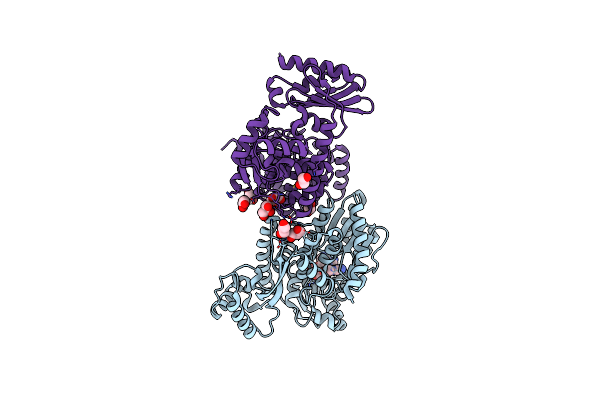 |
The Structure Of Archaeal Nuclease Recj2 From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: IXK, GOL, EDO |
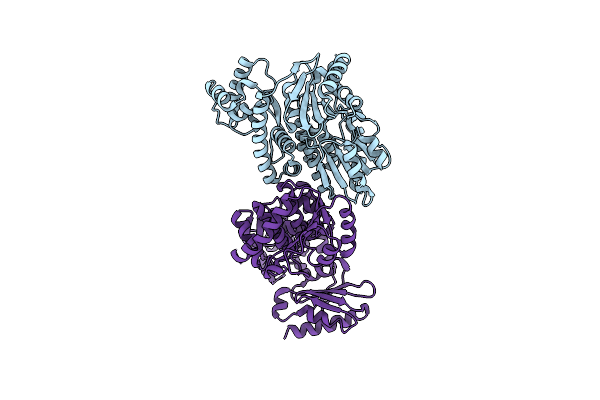 |
The Structure Of Archaeal Nuclease Recj2 From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-07-19 Classification: DNA BINDING PROTEIN |
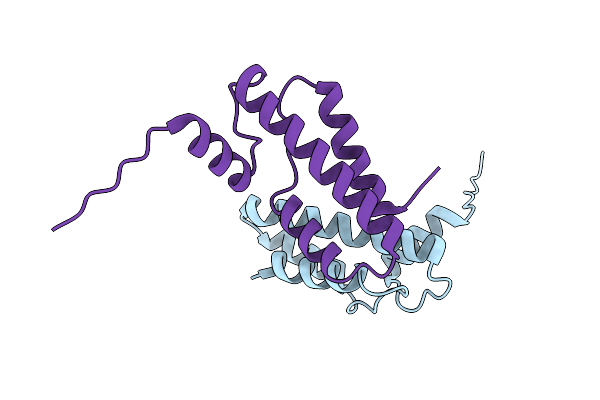 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-07-19 Classification: DNA BINDING PROTEIN |
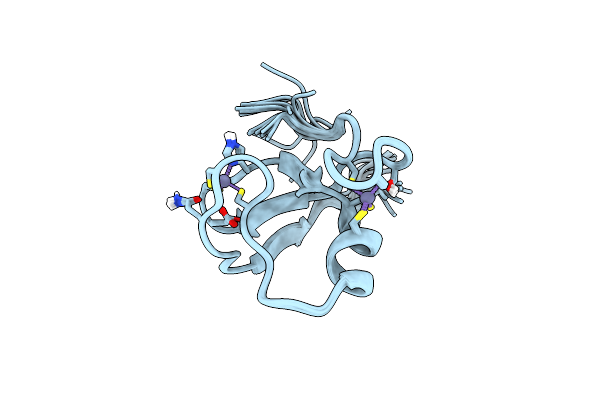 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-07-10 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-14 Classification: HYDROLASE Ligands: MG |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-02-14 Classification: HYDROLASE Ligands: MN, CL |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-02-14 Classification: HYDROLASE Ligands: ZN, CL |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-02-14 Classification: HYDROLASE Ligands: ZN, C5P |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-10-11 Classification: REPLICATION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-10-04 Classification: REPLICATION |
 |
Solution Structure Of Yeast Elongin C In Complex With A Von Hippel-Lindau Peptide
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2001-09-06 Classification: SIGNALING PROTEIN |

