Search Count: 17
 |
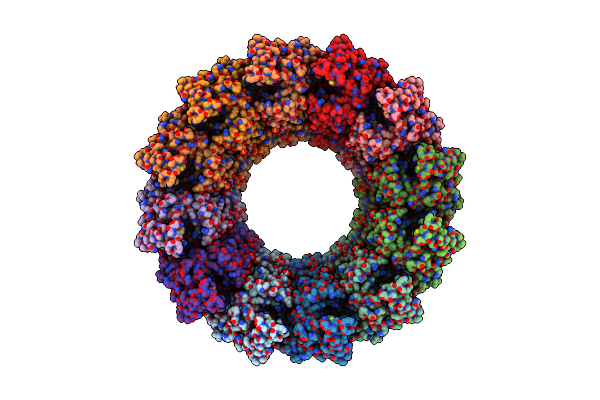
Cryo-Em Structure Of Chikungunya Virus Nonstructural Protein 1 With M7Gpppamu
Organism: Chikungunya virus strain s27-african prototype
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: VIRAL PROTEIN Ligands: ZN, SAH, YG4, MG |
 |
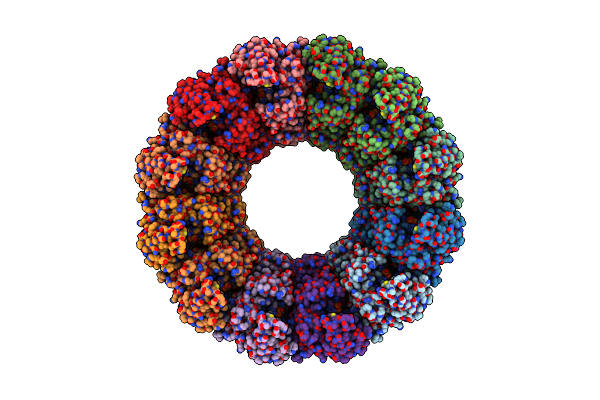
Cryo-Em Structure Of Chikungunya Virus Nonstructural Protein 1 With Inhibitor Fha
Organism: Chikungunya virus strain s27-african prototype
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: VIRAL PROTEIN Ligands: ZN, 7XQ |
 |
Organism: Chikungunya virus strain s27-african prototype
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, MGP, MG |
 |
Organism: Chikungunya virus strain s27-african prototype
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, G7M |
 |
Cryo-Em Structure Of Chikungunya Virus Nonstructural Protein 1 With M7Gppp-Au
Organism: Chikungunya virus strain s27-african prototype, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: VIRAL PROTEIN/RNA Ligands: ZN, SAH, MG |
 |
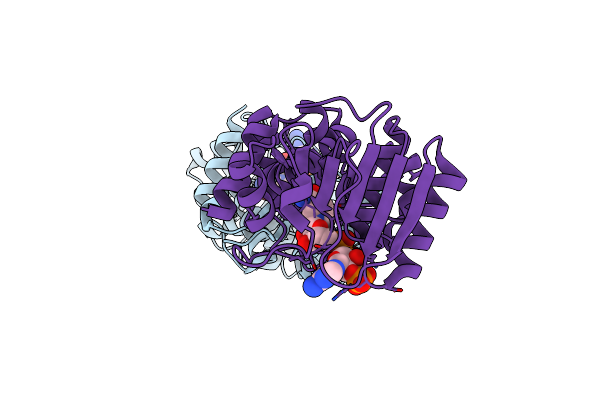
Crystal Structure Of Ligand-Free Form Of 5-Ketofructose Reductase Of Gluconobacter Sp. Strain Chm43
Organism: Gluconobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-08-04 Classification: OXIDOREDUCTASE |
 |
Organism: Gluconobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-08-04 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
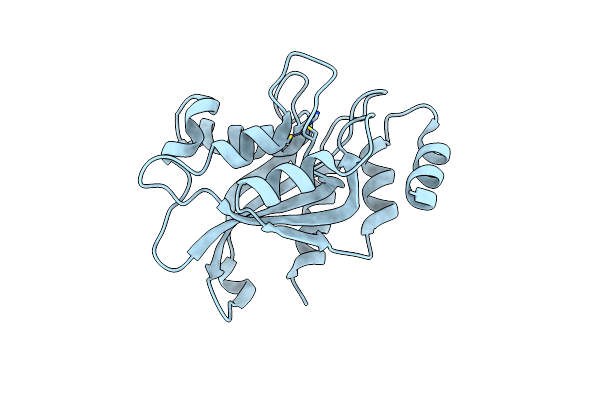
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-07-14 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SO4 |
 |
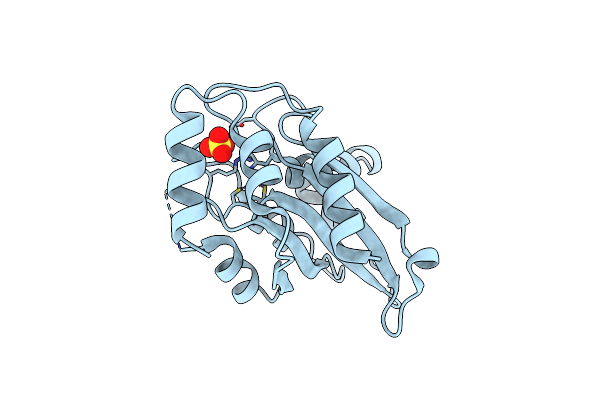
Crystal Structure Of Aspergillus Oryzae Rib2 Deaminase (C-Terminal Deletion Mutant) At Ph 4.6
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
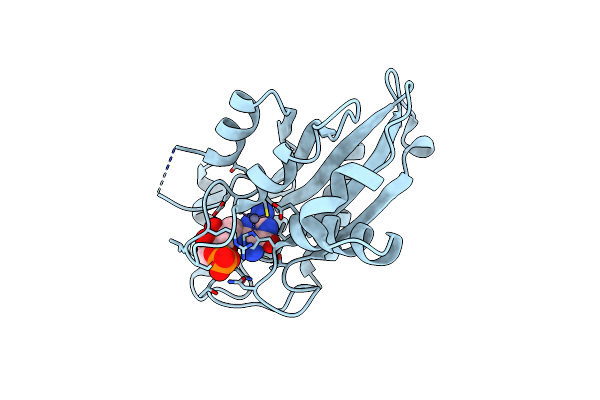
Crystal Structure Of Aspergillus Oryzae Rib2 Deaminase (C-Terminal Deletion Mutant) At Ph 6.5
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-07-14 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SO4 |
 |
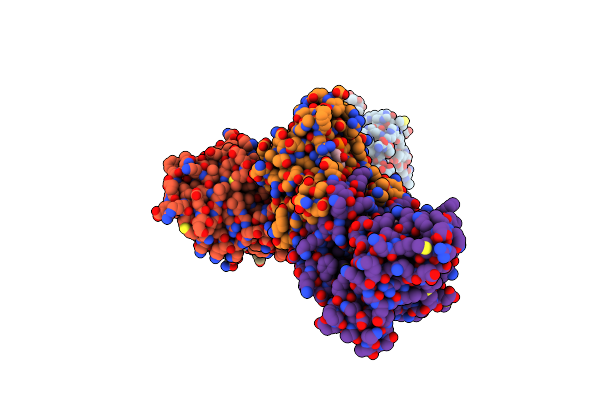
Crystal Structure Of Aspergillus Oryzae Rib2 Deaminase In Complex With Daripp (C-Terminal Deletion Mutant At Ph 6.5)
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-07-14 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, HJL |
 |
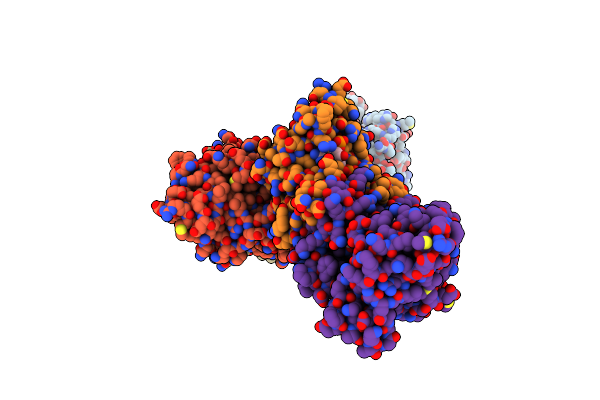
Organism: Methanosarcina mazei (strain atcc baa-159 / dsm 3647 / goe1 / go1 / jcm 11833 / ocm 88)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-07-04 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Methanosarcina mazei (strain atcc baa-159 / dsm 3647 / goe1 / go1 / jcm 11833 / ocm 88)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-07-04 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Methanosarcina mazei (strain atcc baa-159 / dsm 3647 / goe1 / go1 / jcm 11833 / ocm 88)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-04 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Methanosarcina mazei (strain atcc baa-159 / dsm 3647 / goe1 / go1 / jcm 11833 / ocm 88)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-04 Classification: OXIDOREDUCTASE |
 |
Complex Structure Of Bacillus Subtilis Ribg: The Deamination Process In Riboflavin Biosynthesis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2013-02-20 Classification: HYDROLASE, OXIDOREDUCTASE Ligands: ZN, AI9, AOF |
 |
Solution Structure Of The Squash Trypsin Inhibitor Mcoti-Ii, Nmr, 30 Structures.
Organism: Momordica cochinchinensis
Method: SOLUTION NMR Release Date: 2001-04-12 Classification: PROTEASE INHIBITOR |

