Search Count: 37
 |
Organism: Mirabilis jalapa
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: CIT, FE2, TRS, GOL, PEG |
 |
Organism: Epilithonimonas lactis
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN |
 |
Bacterial Sting From Riemerella Anatipestifer In Complex With 3'3'-C-Di-Gmp
Organism: Riemerella anatipestifer yb2
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: C2E, CA |
 |
Organism: Riemerella anatipestifer yb2
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: PG4 |
 |
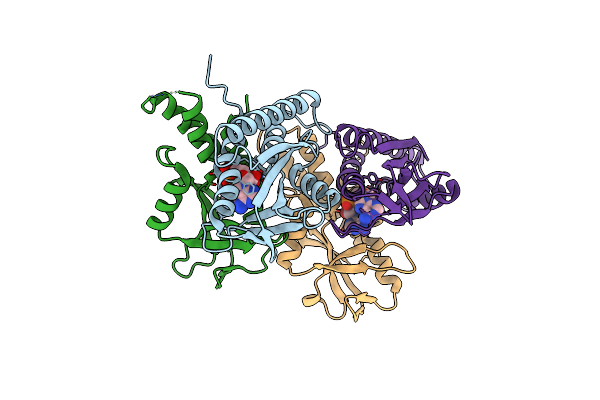
Organism: Larkinella arboricola
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: C2E |
 |
Organism: Epilithonimonas lactis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: 2BA |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C) Varient In Complex With 8-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)Phenoxy)Indolizine-2-Carbonitrile (Jlj555), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN Ligands: 29T |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C) Varient In Complex With 5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)Phenoxy)-2-Naphthonitrile (Jlj600), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN Ligands: 3LQ, MG |
 |
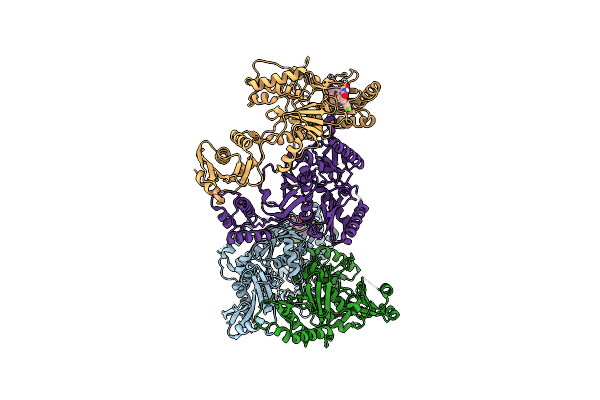
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C) Varient In Complex With 5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)-4-Fluorophenoxy)-7-Fluoro-2-Naphthonitrile (Jlj636), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN Ligands: 7N1 |
 |
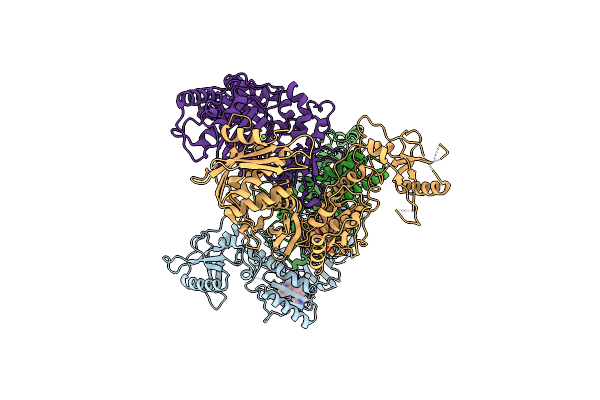
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C, V106A) Varient In Complex With 5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)-4-Fluorophenoxy)-7-Fluoro-2-Naphthonitrile (Jlj636), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus type 1 bh10, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN, Hydrolase Ligands: 7N1, MG |
 |
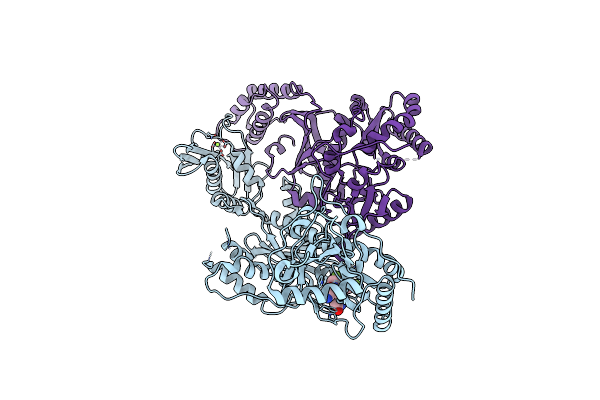
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C, V106A) Varient In Complex With 8-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)Phenoxy)Indolizine-2-Carbonitrile (Jlj555), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus type 1 bh10
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN Ligands: MG, 29T |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C, V106A) Variant In Complex With 8-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)-4-Fluorophenoxy)-6-Fluoroindolizine-2-Carbonitrile (Jlj578), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus type 1 bh10
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN, Hydrolase Ligands: MG, H9Y |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Y181C, V106A) Variant In Complex With 5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)Phenoxy)-2-Naphthonitrile (Jlj600), A Non-Nucleoside Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN, Hydrolase Ligands: SO4, EDO, 3LQ, MG |
 |
Cryo-Em Structure Of The T=4 Lake Sinai Virus 2 Virus-Like Capsid At Ph 7.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=3 Lake Sinai Virus 2 Virus-Like Capsid At Ph 7.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
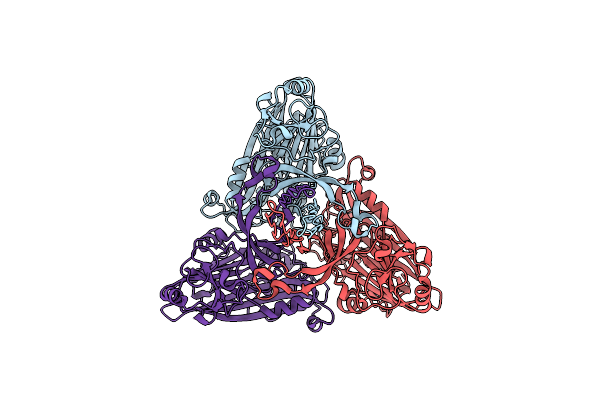
Cryo-Em Structure Of The T=4 Lake Sinai Virus 2 Virus-Like Capsid At Ph 6.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=3 Lake Sinai Virus 2 Virus-Like Capsid At Ph 6.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
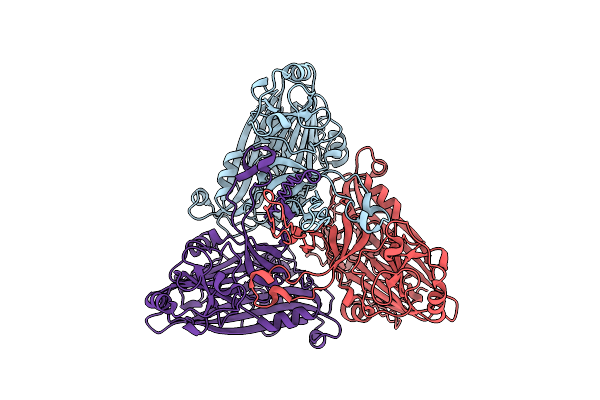
Cryo-Em Structure Of The T=4 Lake Sinai Virus 2 Virus-Like Capsid At Ph 8.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=3 Lake Sinai Virus 2 Virus-Like Capsid At Ph 8.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=3 Lake Sinai Virus 1 (Delta-N48) Virus-Like Capsid At Ph 6.5
Organism: Lake sinai virus 1, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |

