Search Count: 29
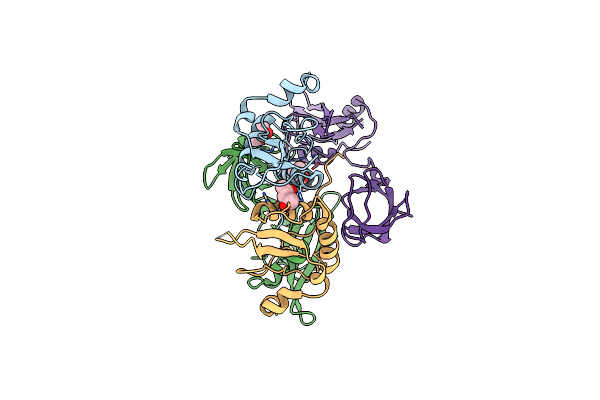 |
Crystal Structure Of Hiv-1 Nef-Sf2 Core Domain In Complex With The Src Family Kinase Hck Sh3-Sh2 Tandem Regulatory Domains
Organism: Human immunodeficiency virus type 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-08-20 Classification: Viral protein/Transferase Ligands: MPD, IOD |
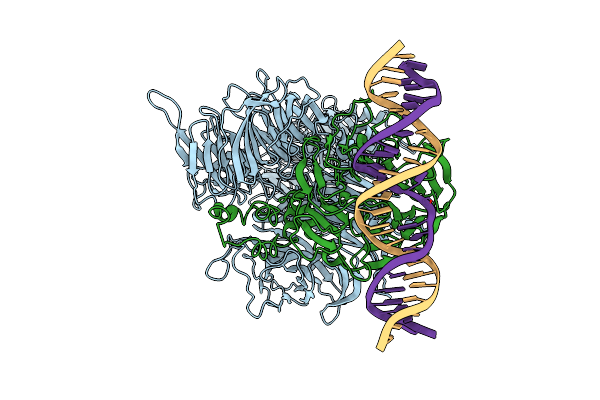 |
Damaged Dna Induced Uv-Damaged Dna-Binding Protein (Uv-Ddb) Dimerization And Its Roles In Chromatinized Dna Repair
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2012-08-22 Classification: DNA BINDING PROTEIN/DNA |
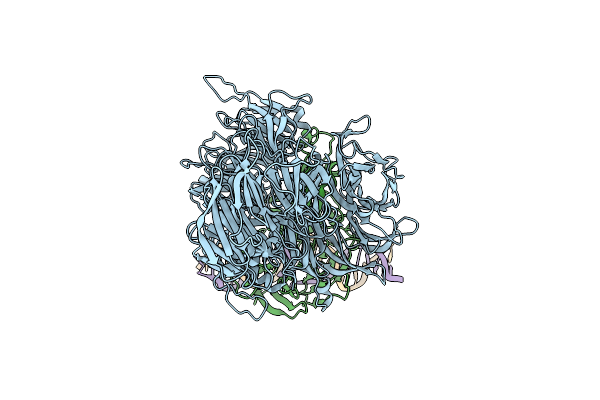 |
Damaged Dna Induced Uv-Damaged Dna-Binding Protein (Uv-Ddb) Dimerization And Its Roles In Chromatinized Dna Repair
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-08-08 Classification: DNA BINDING PROTEIN/DNA |
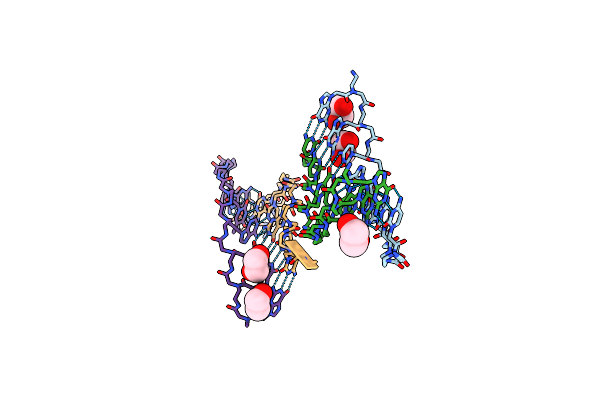 |
Method: X-RAY DIFFRACTION
Resolution:1.27 Å Release Date: 2011-03-30 Classification: Peptide Nucleic Acid Ligands: EDO |
 |
Method: X-RAY DIFFRACTION
Resolution:1.05 Å Release Date: 2011-03-30 Classification: Peptide Nucleic acid Ligands: EDO, PGR, CO3, GOL, PGO |
 |
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2011-02-16 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Chiral Gamma-Pna With Complementary Dna Strand: Insight Into The Stability And Specificity Of Recognition An Conformational Preorganization
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2010-12-29 Classification: PEPTIDE NUCLEIC ACID/DNA Ligands: EDO, ZN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-12-01 Classification: VIRAL PROTEIN Ligands: IOD, I3M, FE, NA |
 |
Crystal Structure Of The Src-Family Kinase Hck Sh3-Sh2-Linker Regulatory Region
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2010-09-01 Classification: TRANSFERASE |
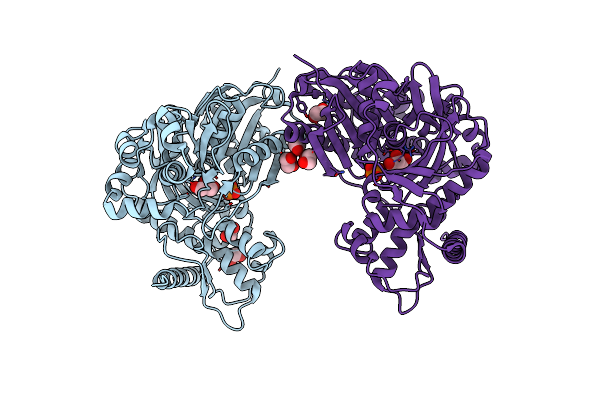 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2009-06-02 Classification: TRANSFERASE Ligands: GOL, EDO, PO4 |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-02 Classification: TRANSFERASE Ligands: EDO, PO4 |
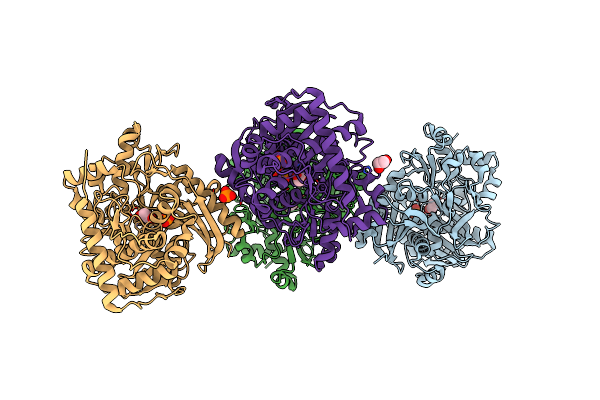 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-06-02 Classification: TRANSFERASE Ligands: EDO, PO4 |
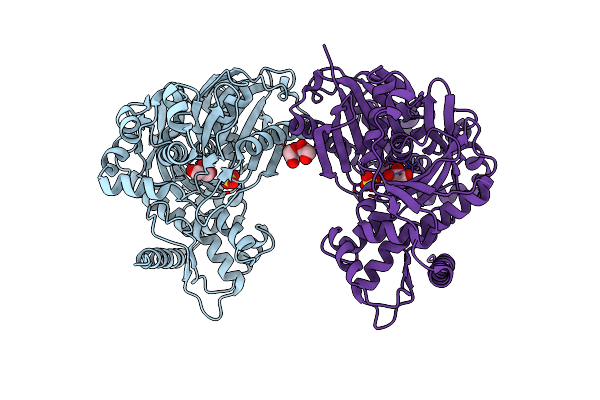 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-06-02 Classification: TRANSFERASE Ligands: SO4, GOL, EDO |
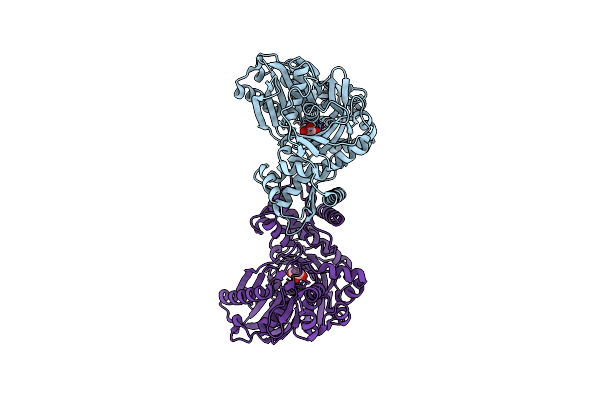 |
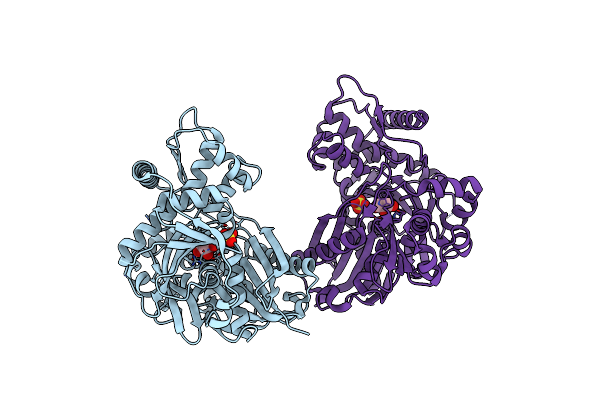
Enterococcus Casseliflavus Glycerol Kinase Mutant His232Ala Complexed With Glycerol
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2009-04-28 Classification: TRANSFERASE Ligands: GOL |
 |
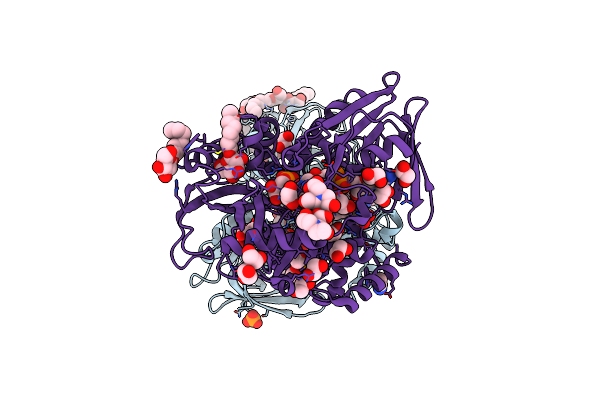
Crystal Structure Of The His-Tagged H232R Mutant Of Glycerol Kinase From Enterococcus Casseliflavus With Glycerol
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-01-20 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
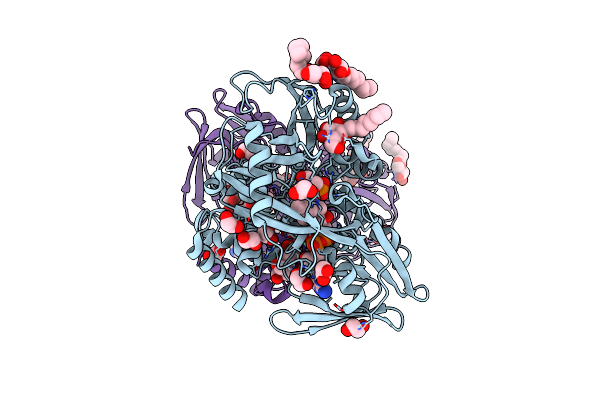
Crystal Structure Of Escherichia Coli Semet Substituted Glycerol-3-Phosphate Dehydrogenase In Complex With Dhap
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2008-06-03 Classification: OXIDOREDUCTASE Ligands: BOG, PO4, FAD, EDO, IMD, 13P, BCN, TAM |
 |
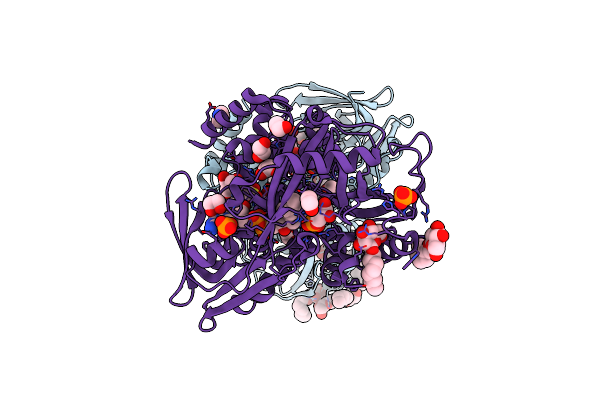
Crystal Structure Of Escherichia Coli Glycerol-3-Phosphate Dehydrogenase In Complex With 2-Phosphopyruvic Acid.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-29 Classification: OXIDOREDUCTASE Ligands: BOG, EDO, T3A, FAD, PEP, IMD |
 |
Crystal Structure Of Escherichia Coli Glycerol-3-Phosphate Dehydrogenase In Complex With Dhap
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-29 Classification: OXIDOREDUCTASE Ligands: BOG, PO4, FAD, 13P, EDO, IMD, T3A |
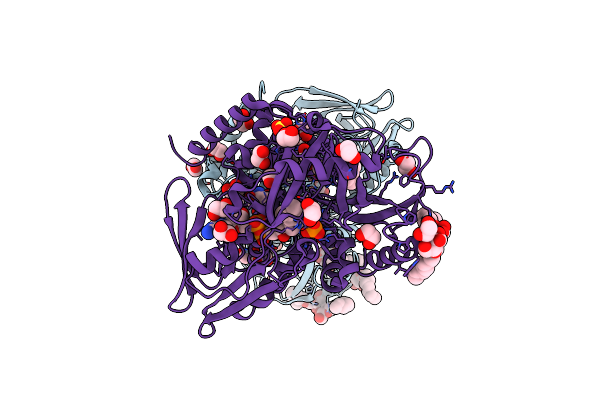 |
Crystal Structure Of Glycerol-3-Phosphate Dehydrogenase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2008-04-15 Classification: OXIDOREDUCTASE Ligands: BOG, SO4, PO4, FAD, TAM, EDO, IMD |
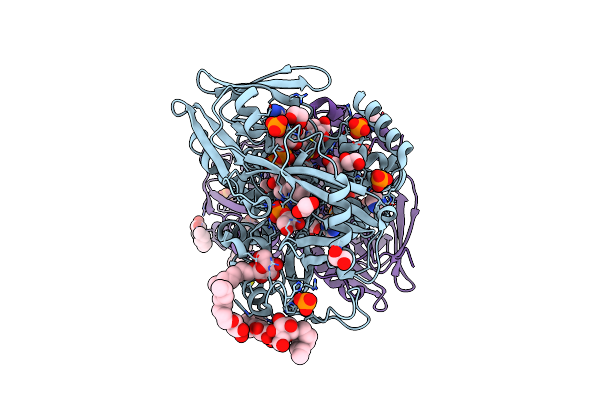 |
Crystal Structure Of Escherichia Coli Glycerol-3-Phosphate Dehydrogenase In Complex With 2-Phospho-D-Glyceric Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-15 Classification: OXIDOREDUCTASE Ligands: BOG, PO4, FAD, IMD, EDO, T3A, 2PG |

