Search Count: 24
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-11-20 Classification: DE NOVO PROTEIN Ligands: HCY, SO4 |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-06-10 Classification: HYDROLASE Ligands: MES, UKV |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-06-10 Classification: HYDROLASE/INHIBITOR Ligands: UL1, MG, MES |
 |
Crystal Structure Of Disulfide Bond Oxidoreductase Dsba1 From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-04-24 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Crystal Structure Of Putative Sufu (Suppressor Of Fused Protein) Homolog (Yp_208451.1) From Neisseria Gonorrhoeae Fa 1090 At 1.40 A Resolution
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-26 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Crystal Structure Of Putative Cell Invasion Protein With Mac/Perforin Domain (Np_812351.1) From Bacteriodes Thetaiotaomicron Vpi-5482 At 2.46 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2009-11-24 Classification: CELL INVASION Ligands: CL, EDO, MPD |
 |
Crystal Structure Of A Putative Glycoside Hydrolase (Bt_2081) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.05 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-05-26 Classification: Ca-BINDING PROTEIN Ligands: CA, NA, CAC, ACT, PEG, P6G |
 |
Crystal Structure Of A Nlpc/P60 Family Protein (Bce_2878) From Bacillus Cereus Atcc 10987 At 1.79 A Resolution
Organism: Bacillus cereus atcc 10987
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: ALA, DGL, PO4, PG4 |
 |
Crystal Structure Of Putative Polysaccharide Binding Proteins (Duf1812) (Np_809975.1) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.20 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-24 Classification: Carbohydrate Binding Protein Ligands: EDO |
 |
Crystal Structure Of Glycoprotein Endopeptidase (Tm0874) From Thermotoga Maritima At 2.50 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-07-19 Classification: HYDROLASE Ligands: UNL |
 |
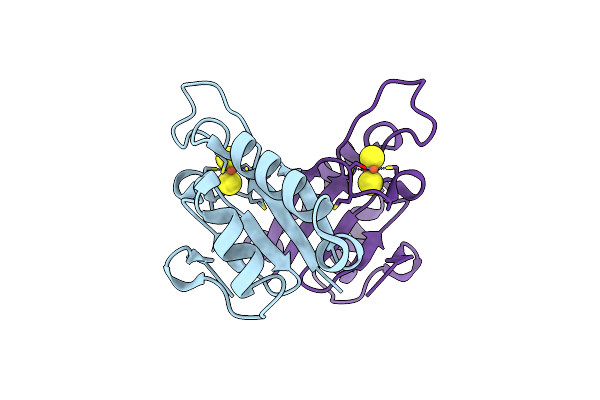
Crystal Structure At 1.5 Angstroms Resolution Of The Wild Type Thioredoxin-Like [2Fe-2S] Ferredoxin From Aquifex Aeolicus
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-09-18 Classification: ELECTRON TRANSPORT Ligands: ZN, FES, SO4 |
 |
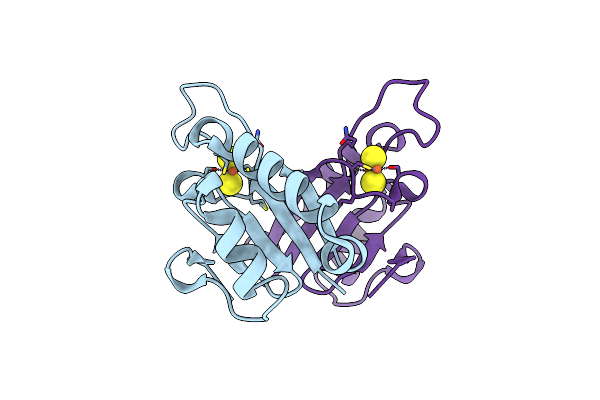
Crystal Structure At 1.25 Angstroms Resolution Of The Cys55Ser Variant Of The Thioredoxin-Like [2Fe-2S] Ferredoxin From Aquifex Aeolicus
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2002-09-18 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Crystal Structure At 1.05 Angstroms Resolution Of The Cys59Ser Variant Of The Thioredoxin-Like [2Fe-2S] Ferredoxin From Aquifex Aeolicus
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2002-09-18 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
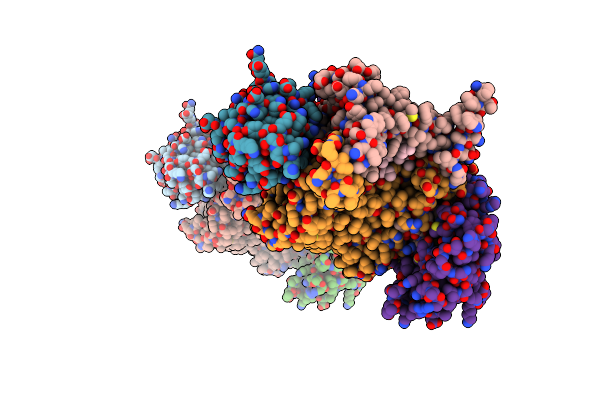
X-Ray Structure Of The Cytochrome-C(2)-Photosynthetic Reaction Center Electron Transfer Complex From Rhodobacter Sphaeroides In Type Ii Co-Crystals
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-06-12 Classification: PHOTOSYNTHESIS |
 |
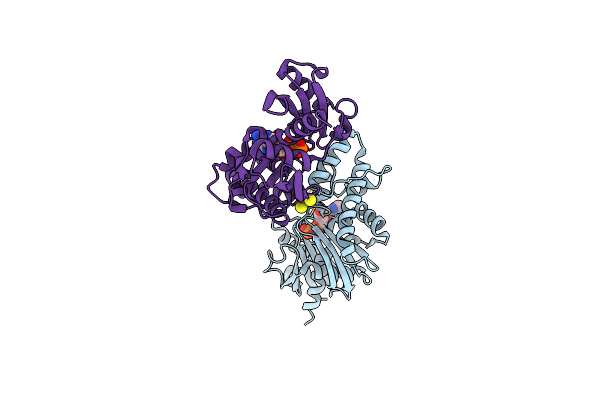
X-Ray Structure Of The Cytochrome-C(2)-Photosynthetic Reaction Center Electron Transfer Complex From Rhodobacter Sphaeroides In Type I Co-Crystals
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2002-06-12 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, FE2, CL, U10, LDA, HEM |
 |
Crystal Structure Of The Acidaminococcus Fermentans (R)-2-Hydroxyglutaryl-Coa Dehydratase Component A
Organism: Acidaminococcus fermentans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2001-03-21 Classification: METAL BINDING PROTEIN Ligands: SF4, ADP |

