Search Count: 353
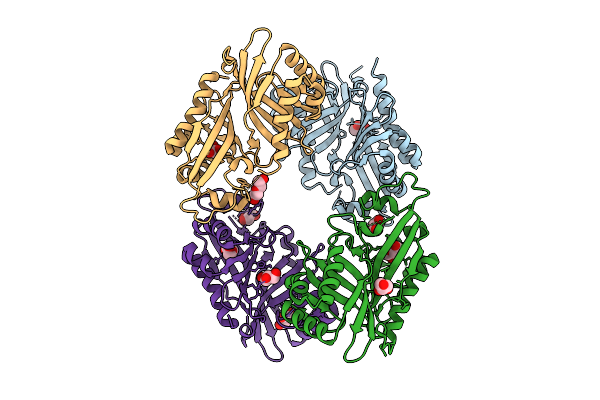 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
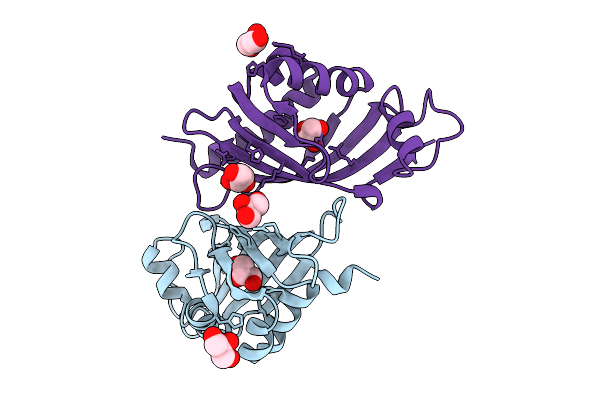 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
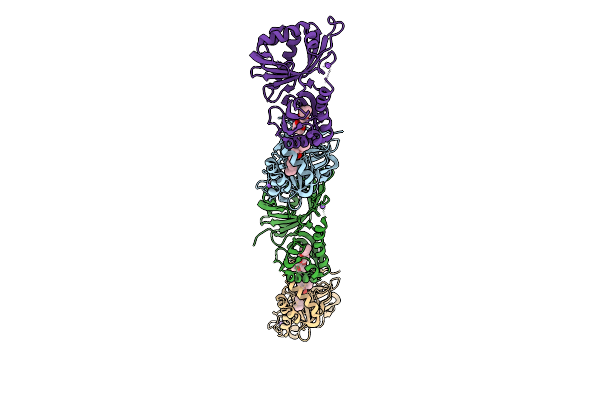 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
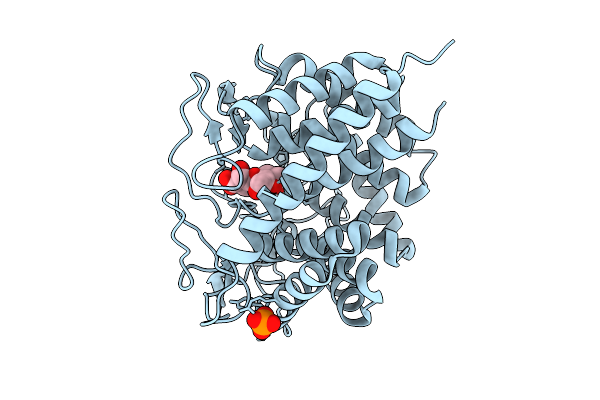 |
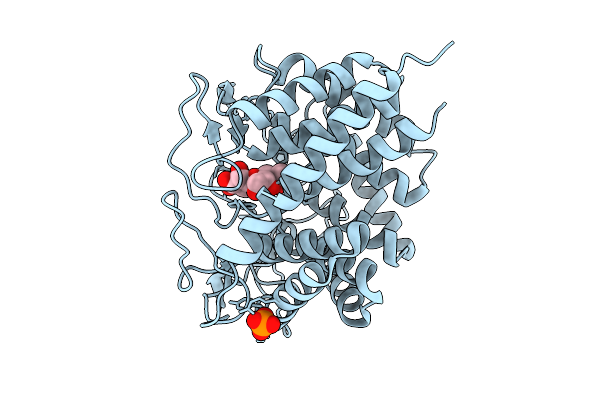
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiose
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiitol
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: MTL, BMA, CL, PO4 |
 |
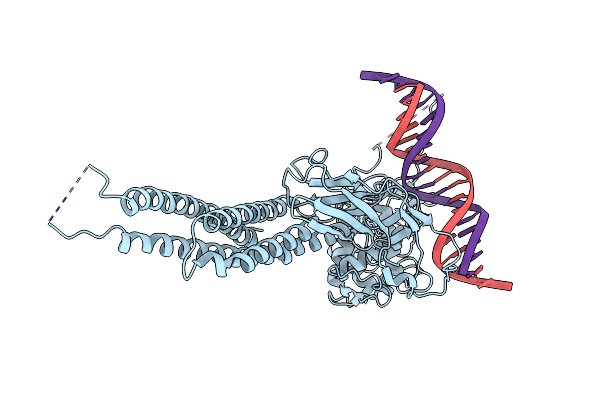
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2025-03-26 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Tyrosine Decarboxylase H203F Mutant In Complex With The Cofactor Plp And Tyrosine
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: TYR |
 |
Crystal Structure Of Tyrosine Decarboxylase In Complex With The Cofactor Plp And Inhibitor Carbidopa
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: PLP, 142 |
 |
Crystal Structure Of Gh13_30 Alpha-Glucosidase Cmmb In Complex With Acarbose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-07-03 Classification: HYDROLASE |
 |
Crystal Structure Of Gh13_30 Alpha-Glucosidase Cmmb In Complex With Acarviosin
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: A1L2I |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2024-05-29 Classification: REPLICATION |
 |
Organism: Callicarpa macrophylla
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: EDO |
 |
Glycoside Hydrolase Family 55 Endo-Beta-1,3-Glucanase From Microdochium Nivale
Organism: Microdochium nivale
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GOL, ACE |
 |
Eisenia Hydrolysis-Enhancing Protein From Aplysia Kurodai Complex With Tannic Acid
Organism: Aplysia kurodai
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-15 Classification: UNKNOWN FUNCTION Ligands: GGP |
 |
Organism: Indibacter alkaliphilus
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-11-01 Classification: METAL TRANSPORT Ligands: RET |
 |
Organism: Theaceae
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2023-09-20 Classification: TRANSFERASE Ligands: SAH, MG |

