Search Count: 47
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Ligand-Free Form At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Glucose Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: BGC, NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE, DOD |
 |
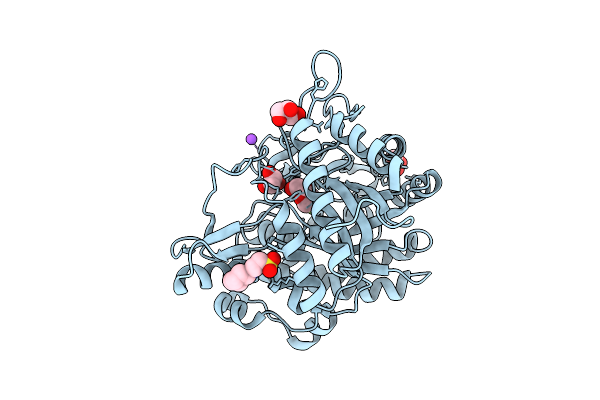
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE |
 |
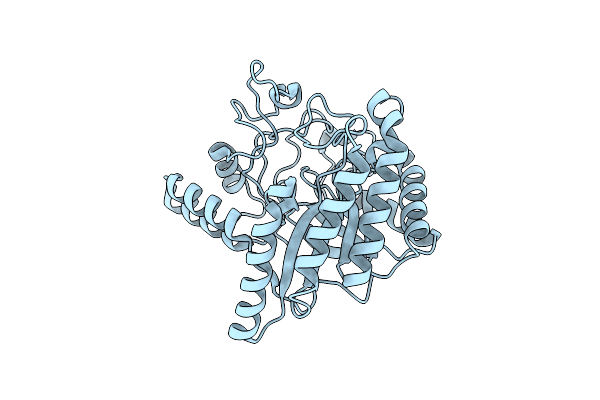
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Cryogenic Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NA, GOL, NHE |
 |
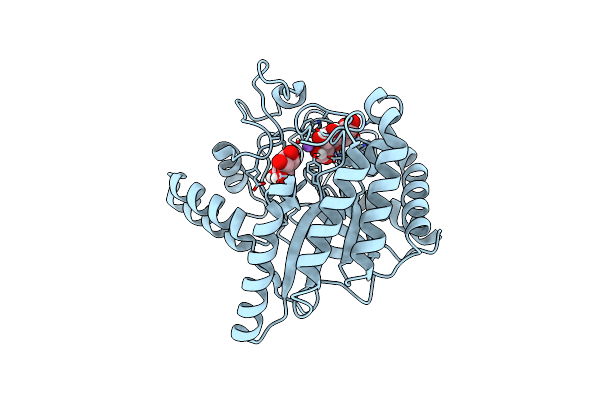
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.36 Å, 1.86 Å Release Date: 2025-03-12 Classification: HYDROLASE |
 |
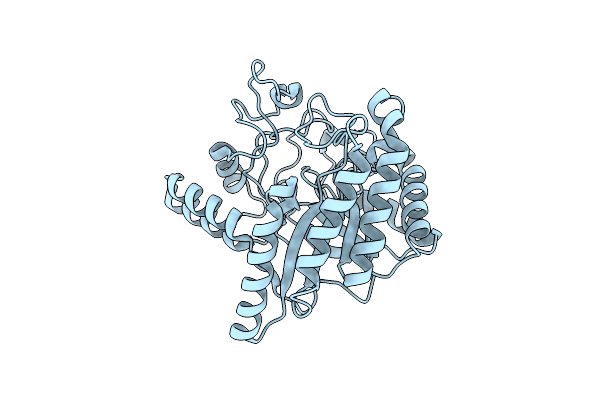
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 1.8 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Low-D2O-Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 2.15 Å Release Date: 2025-03-12 Classification: HYDROLASE |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex, H2O Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.80 Å, 2.59 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: MPD, ACT |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA, CL, PEG |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Reduced Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Oxidized Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, GOL, FAD |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Reduced Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
R115A Mutant Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
The Structure Of Ente With 3-(Prop-2-Yn-1-Yloxy)Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VPT |
 |
The Structure Of Ente With2-Methyl-3-Chloro-Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VQ5 |
 |
Neutron Structure Of Bacillus Thermoproteolyticus Ferredoxin At Room Temperature
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.45 Å, 1.60 Å Release Date: 2024-02-07 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
X-Ray Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex At 100K
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2024-01-24 Classification: LYASE Ligands: RAM, ACT, TRS, CA, NA |

