Search Count: 24
 |
Organism: Pseudomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Single-Chain L-Glutamate Oxidase Mutant From Streptomyces Sp. X-119-6
Organism: Streptomyces sp. x-119-6
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2023-08-09 Classification: OXIDOREDUCTASE Ligands: FAD, AKG, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of L-Histidine Decarboxylase (C57S Mutant) From Photobacterium Phosphoreum
Organism: Photobacterium phosphoreum
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-02-16 Classification: LYASE |
 |
Crystal Structure Of L-Histidine Decarboxylase (C57S/C101V/C282V Mutant) From Photobacterium Phosphoreum
Organism: Photobacterium phosphoreum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-02-16 Classification: LYASE Ligands: IMD |
 |
Ambient Temperature Structure Of Bifidobacterium Longum Phosphoketolase With Thiamine Diphosphate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: TPP, CA, LMR, MLA, SIN |
 |
Ambient Temperature Structure Of Bifidobacgterium Longum Phosphoketolase With Thiamine Diphosphate And Phosphoenol Pyuruvate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: PEP, TPP, CA |
 |
Organism: Bifidobacterium longum subsp. longum f8
Method: ELECTRON MICROSCOPY Release Date: 2021-02-17 Classification: LYASE Ligands: TPP, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL |
 |
Crystal Structure Of Fabp4 In Complex With 3-(5-Cyclopropyl-2,3-Diphenyl-1H-Indol-1-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: 57P |
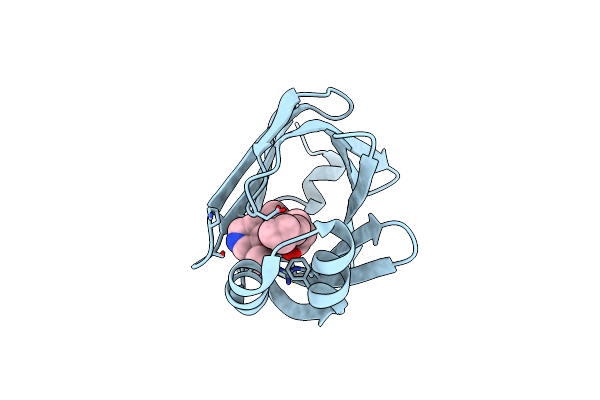 |
Crystal Structure Of Fabp4 In Complex With 3-[5-Cyclopropyl-3-(3-Methoxypyridin-4-Yl)-2-Phenyl-1H-Indol-1-Yl] Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: L19 |
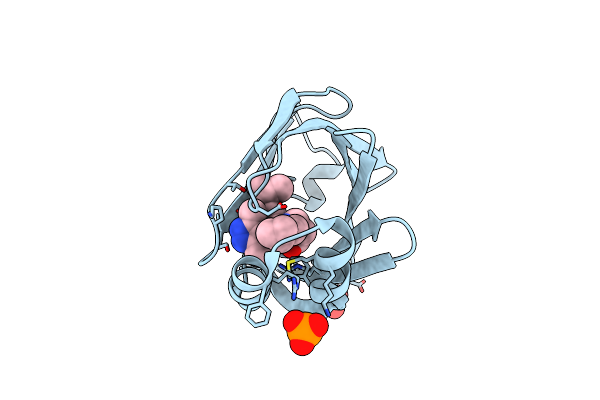 |
Crystal Structure Of Fabp4 In Complex With 3-{5-Cyclopropyl-3-(3,5-Dimethyl-1H-Pyrazol-4-Yl)-2-[3-(Propan-2-Yloxy) Phenyl]-1H-Indol-1-Yl}Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: L96, PO4 |
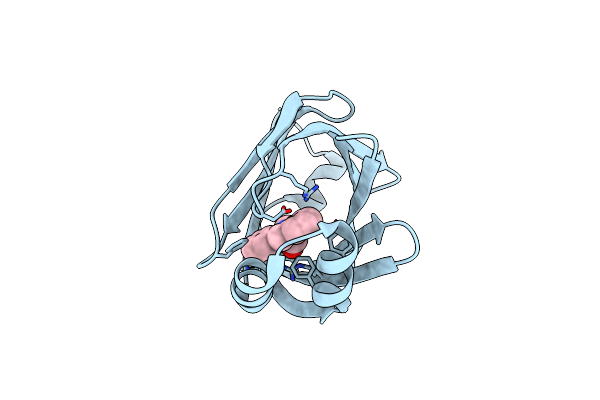 |
Crystal Structure Of Fabp4 In Complex With 3-(2-Phenyl-1H-Indol-1-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: LIPID BINDING PROTEIN Ligands: 57Q |
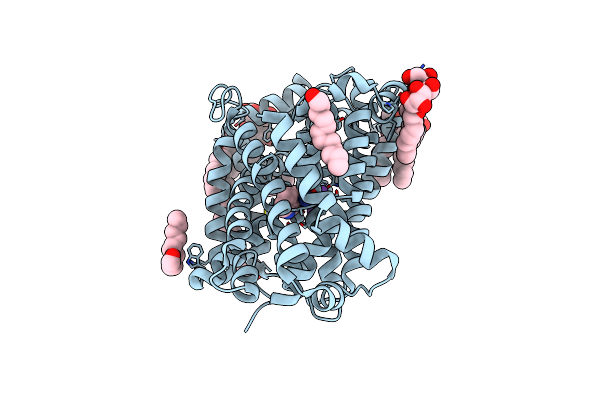 |
Crystal Structure Of The Bacterial Nss Member Mhst In An Occluded Inward-Facing State
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN Ligands: TRP, NA, BNG, LMU |
 |
Crystal Structure Of The Bacterial Nss Member Mhst In An Occluded Inward-Facing State (Lipidic Cubic Phase Form)
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN Ligands: TRP, NA, 78M, 78N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-11-06 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, HPU, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-11-06 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, HPW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2012-12-12 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, EPE, HPJ |

