Search Count: 698
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: CA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
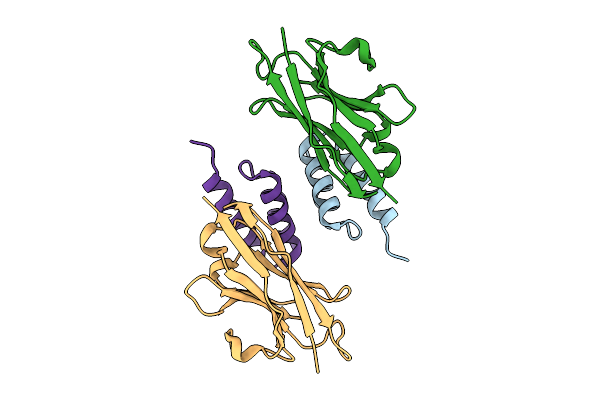 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
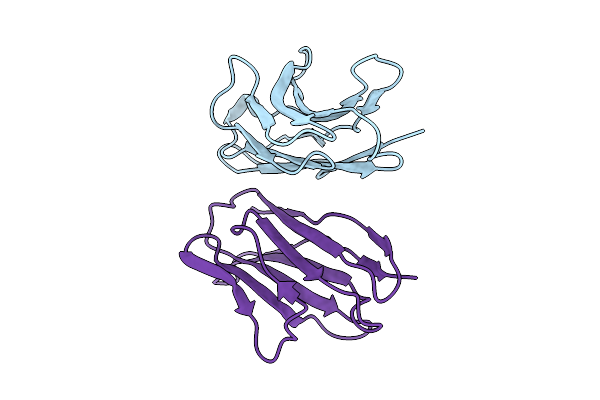 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM |
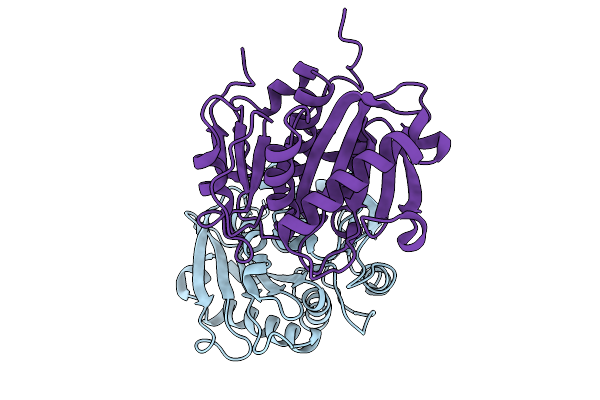 |
Organism: Bacterium hr29
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE |
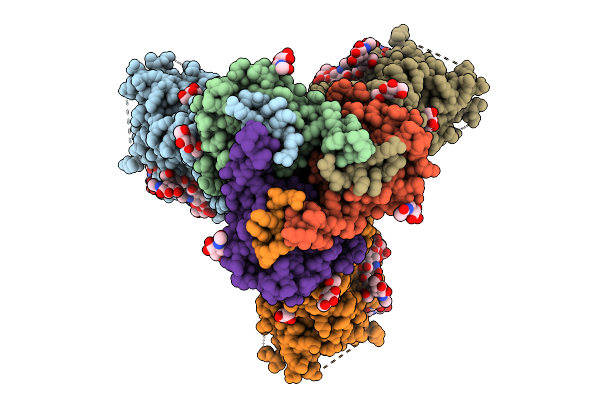 |
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Importin Alpha Isoform 2 With Synthetic Zero Net-Charge Nuclear Localization Signal
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: GOL, DTT |
 |
Importin Alpha Isoform 2 With Synthetic Zero Net-Charge Nuclear Localization Signal
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: GOL, DTT |
 |
Importin Alpha Isoform 2 With Synthetic Zero Net-Charge Nuclear Localization Signal
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: GOL, DTT |
 |
Importin Alpha Isoform 2 With Synthetic Zero Net-Charge Nuclear Localization Signal
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: GOL, DTT |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DNA BINDING PROTEIN |
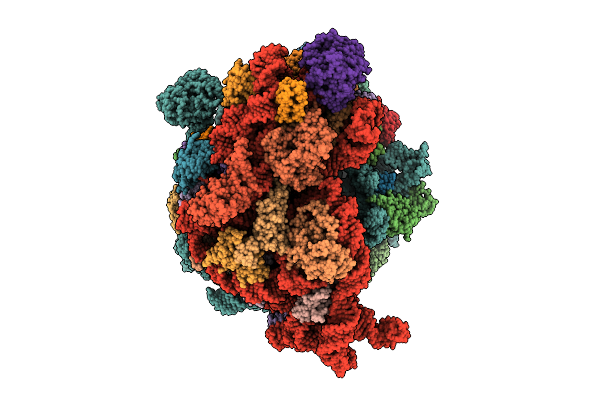 |
Cryo-Em Structure Of Escherichia Coli Hibernating Ribosome With Rnase I Mutant
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RIBOSOME Ligands: MG, CA |
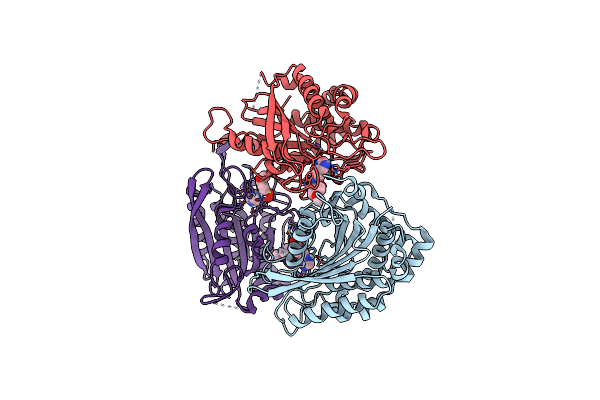 |
Crystal Structure Of Human Glutaminyl Cyclase In Complex With N-(2-(1H-Imidazol-5-Yl)Ethyl)-4-Methoxybenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: ZN, A1EFY |

