Search Count: 1,067
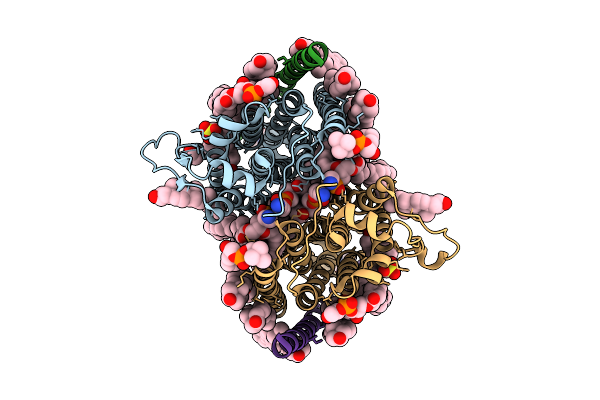 |
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta Bound With Tlca
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: CLR, P0E, LPE, NAG, PLM, A1EPX, 76F |
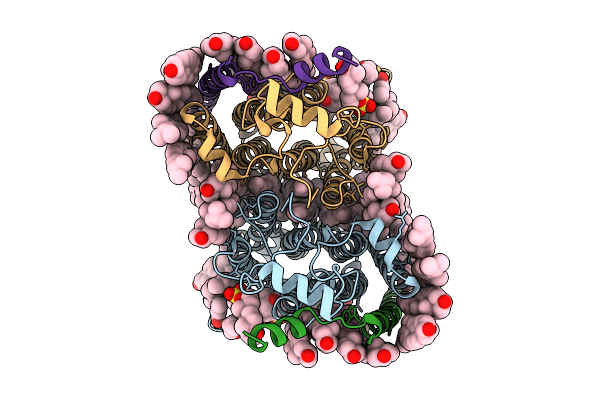 |
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta Bound With Dheas
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: 76F, P0E, LPE, CLR, PLM, ZWY |
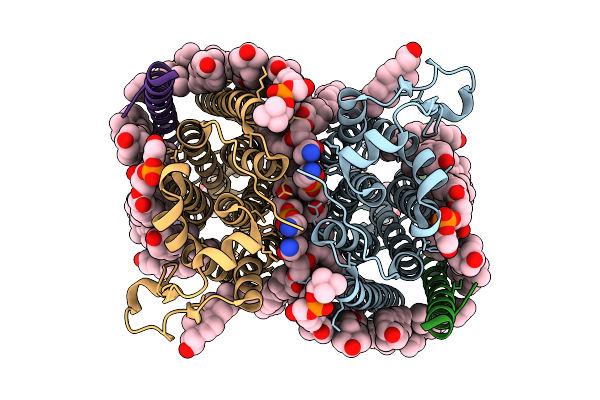 |
Cryo-Em Structure Of Human Organic Solute Transporter Ost-Alpha/Beta In Apo State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: 76F, P0E, LPE, CLR, NAG, PLM |
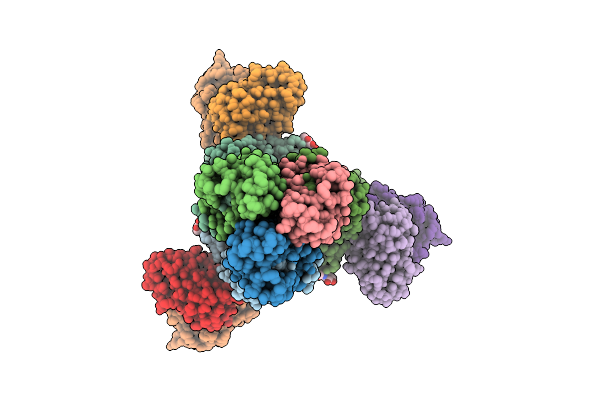 |
Cryoem Structure Of H7 Hemagglutinin In Complex With A Human Neutralizing Antibody 6Y13
Organism: Influenza a virus (a/duck/chiba/25-51-14/2013(h7n1)), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: NAG |
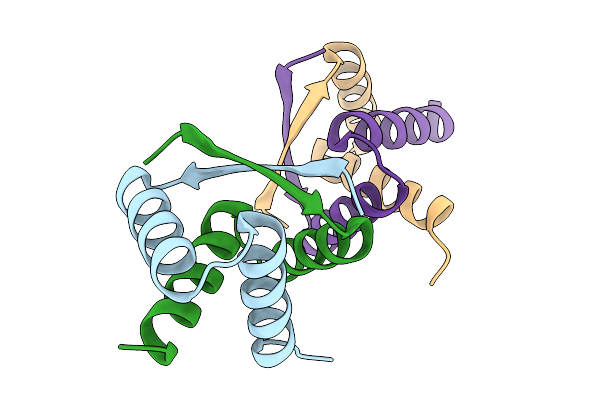 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
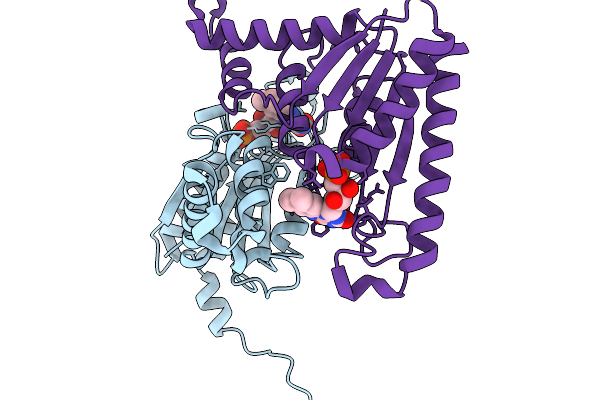 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
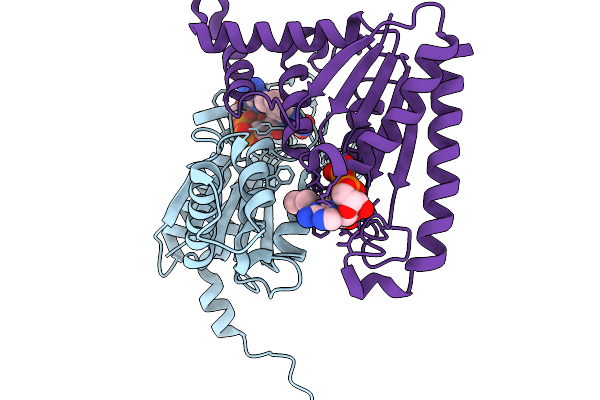 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
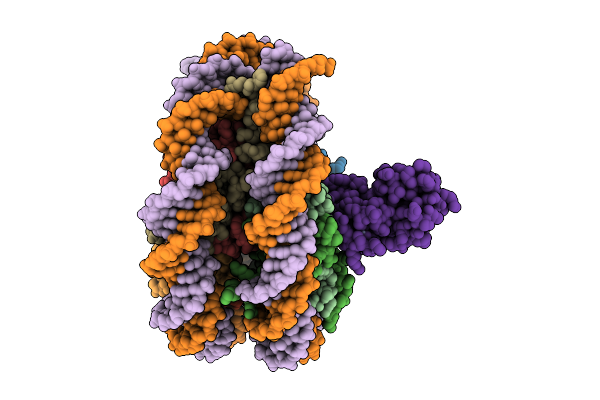 |
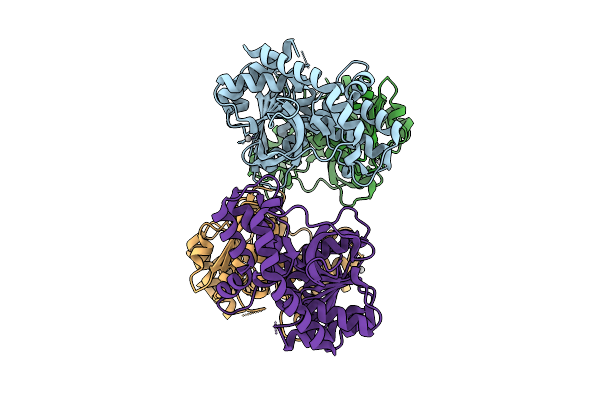
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
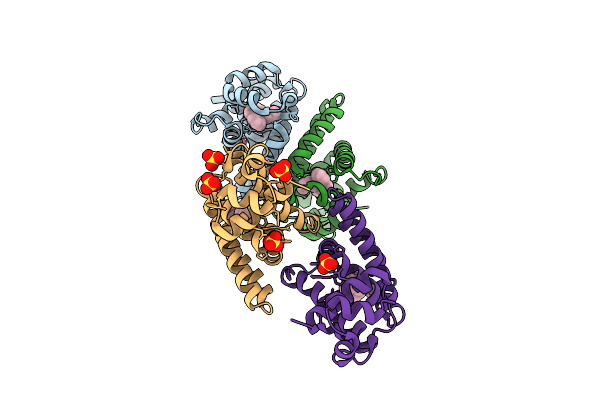
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOSOLIC PROTEIN Ligands: AG |
 |
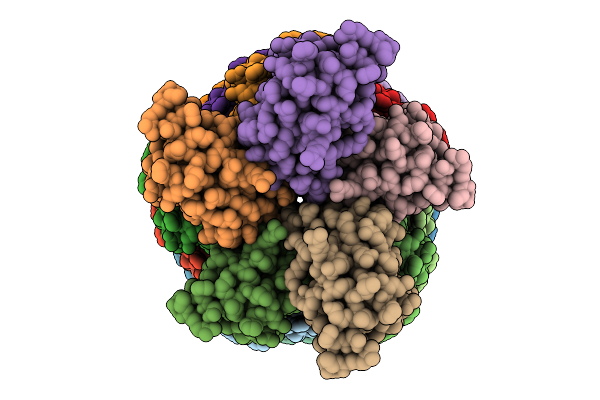
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: A1EKZ, SO4 |
 |
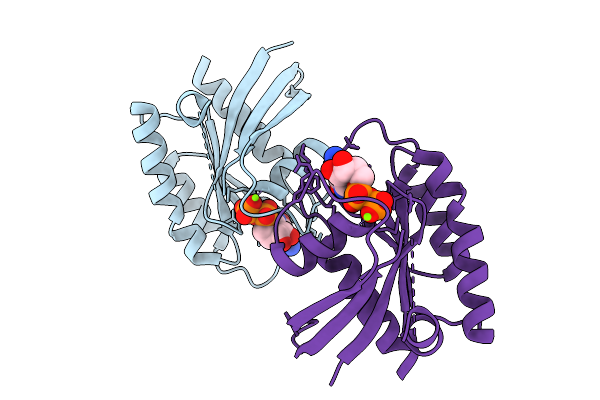
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: IMMUNE SYSTEM |
 |
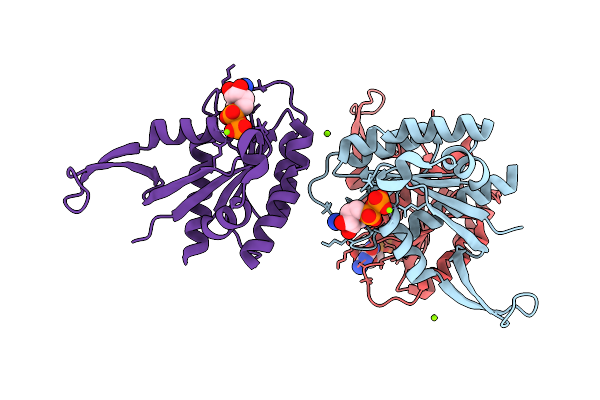
Crystal Structure Of Gdp-Bound Kras G12D/M67R: Suppressing G12D Oncogenicity Via Second-Site M67R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
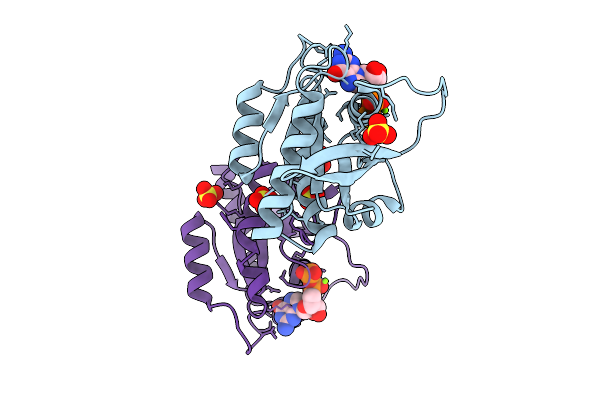
Crystal Structure Of Gdp-Bound Kras G12D/I55E: Suppressing G12D Oncogenicity Via Second-Site I55E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
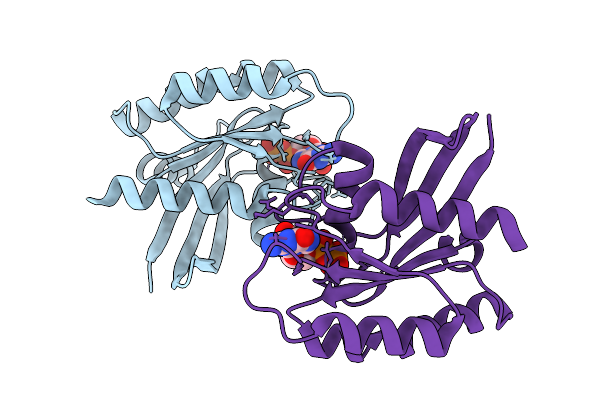
Crystal Structure Of Gdp-Bound Kras G12D/F28K: Suppressing G12D Oncogenicity Via Second-Site F28K Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
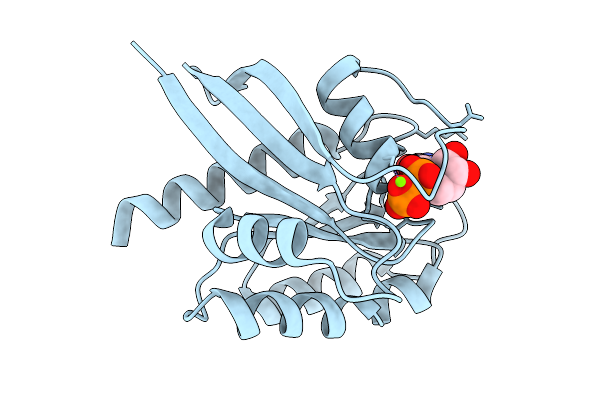
Crystal Structure Of Gdp-Bound Kras G12D/P34R: Suppressing G12D Oncogenicity Via Second-Site P34R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
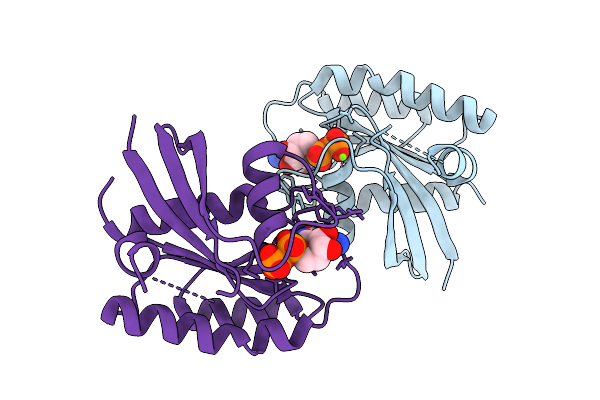
Crystal Structure Of Gdp-Bound Kras G12D/R41Q: Suppressing G12D Oncogenicity Via Second-Site R41Q Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
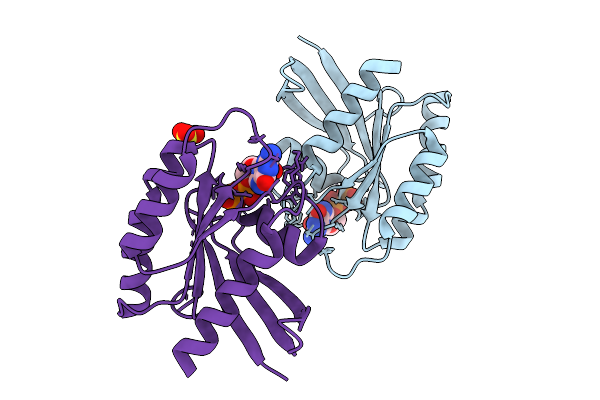
Crystal Structure Of Gdp-Bound Kras G12D/V45E: Suppressing G12D Oncogenicity Via Second-Site V45E Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
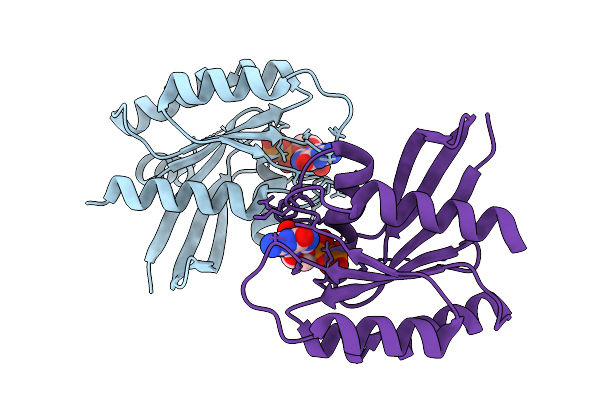
Crystal Structure Of Gdp-Bound Kras G12D/D54R: Suppressing G12D Oncogenicity Via Second-Site D54R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: GDP, MG, SO4 |
 |
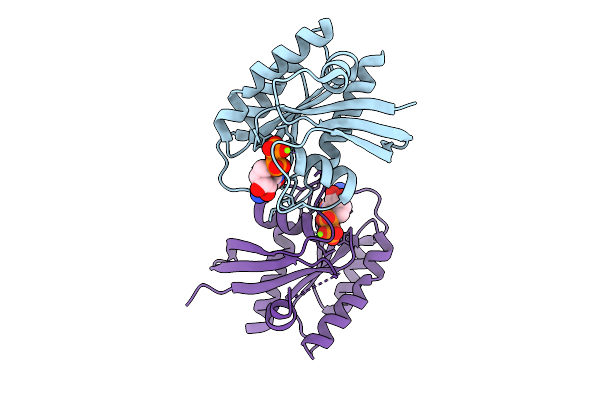
Crystal Structure Of Gdp-Bound Kras G12D/G60R: Suppressing G12D Oncogenicity Via Second-Site G60R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
Crystal Structure Of Gdp-Bound Kras G12D/V103Y: Suppressing G12D Oncogenicity Via Second-Site V103Y Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ONCOPROTEIN Ligands: MG, GDP |

