Search Count: 680
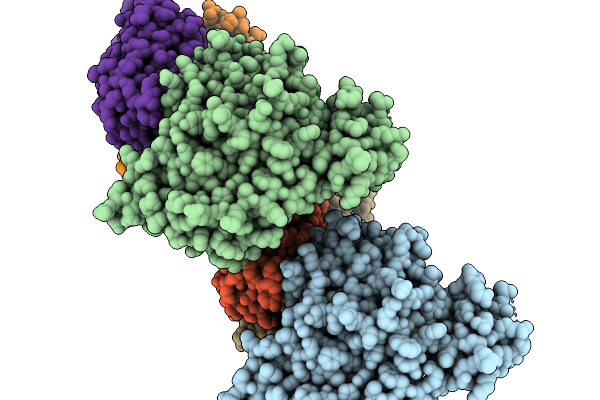 |
Organism: Severe fever with thrombocytopenia syndrome virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
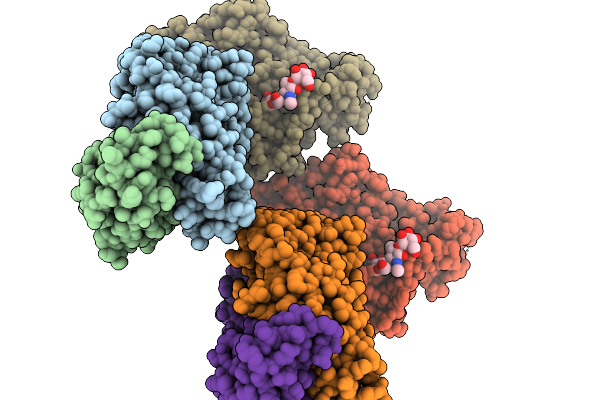 |
Organism: Homo sapiens, Severe fever with thrombocytopenia syndrome virus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
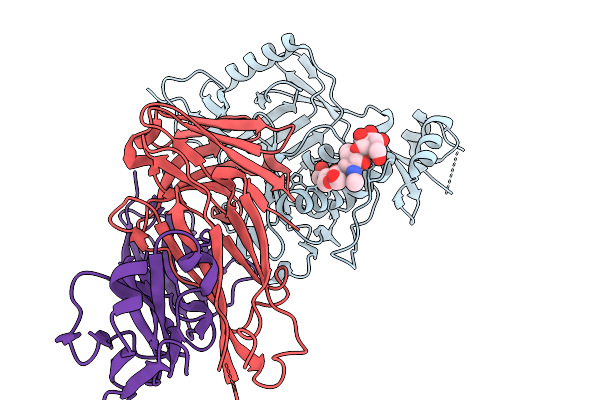 |
Organism: Homo sapiens, Severe fever with thrombocytopenia syndrome virus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
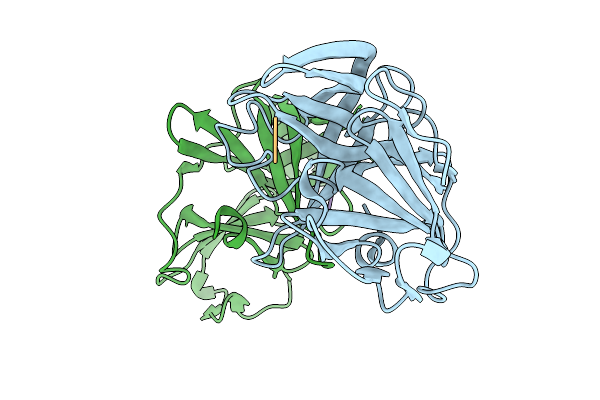 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Crystal Structure Of Sars-Cov-2 Papain-Like Protease (Cys111Ser) In Complex With Yl1004
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EOE, ZN, CL, PEG |
 |
Omicron-Specific Ultra-Potent Sars-Cov-2 Neutralizing Antibodies Targeting The N1/N2 Loop Of Spike N-Terminal Domain
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: A1ELW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: GSH, FBP |
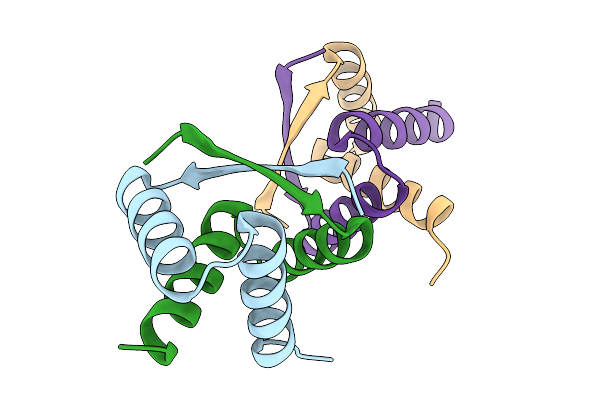 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
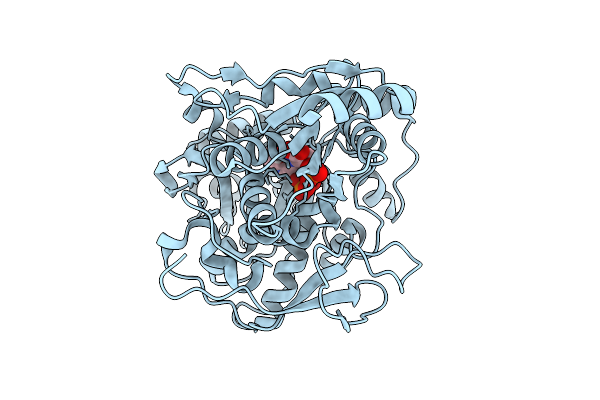 |
Crystal Structure Of Capa2 From Alistipes Finegoldii In Complex With Ops-Plp External Aldimine
Organism: Alistipes finegoldii dsm 17242
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: E1U |
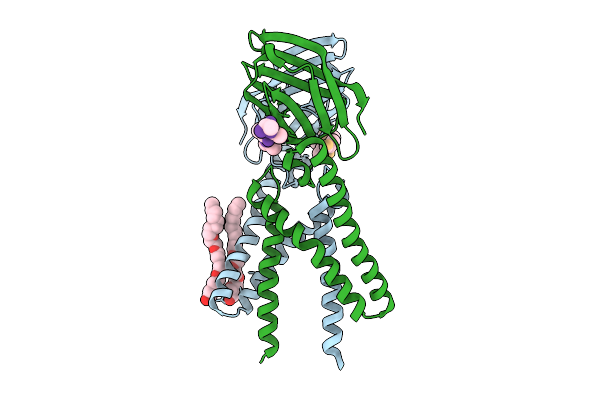 |
Crystal Structure Of A Coronaviral M Protein In Complex With A C-Terminal Peptide Of The N Protein
Organism: Pipistrellus bat coronavirus hku5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: N8E |
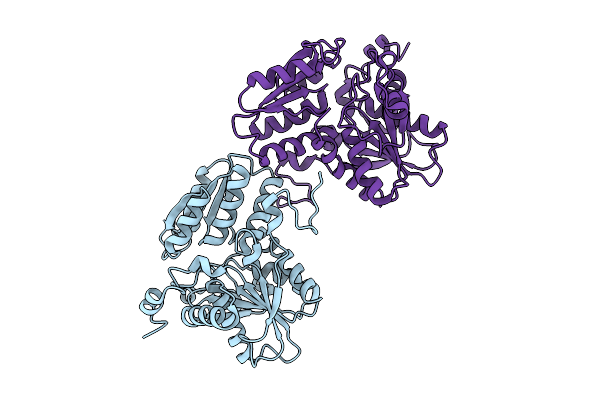 |
Organism: Capnocytophaga ochracea dsm 7271
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Capa2 From Alistipes Finegoldii In Complex With Plp Cofactor
Organism: Alistipes finegoldii dsm 17242
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, PLP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Sars-Cov-2 Omicron Main Protease (Mpro) Complex With Azapeptide Inhibitor 20A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: A1BKV |
 |
Nmr Rdc Refinement Of The Helical Domain Of The Sars-Cov-2 Monomeric Main Protease (Mproh41Q,10-306)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: HYDROLASE |
 |
Nmr Rdc Refinement Of The Catalytic Domain Of The Sars-Cov-2 Monomeric Main Protease (Mproh41Q,10-306)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: HYDROLASE |
 |
Crystal Structure Of The Moae-Like Domain Within Rv3323C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: MES, GOL |

