Search Count: 530
 |
Organism: Homo sapiens
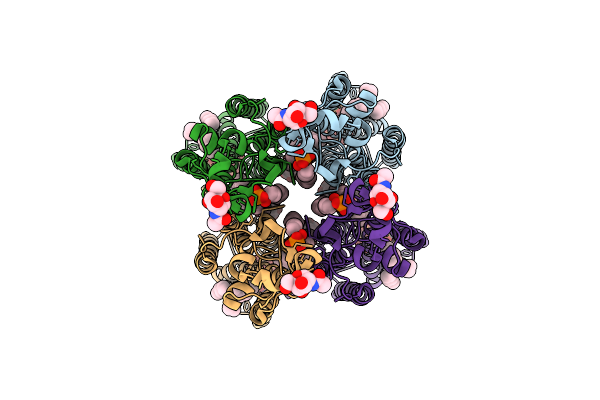
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LPP, NAG, LBN, PT5 |
 |
Cryo-Em Structure Of Human Lipid Phosphate Phosphatase 1 Complexed With Lpa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: NAG, LBN, PT5, NKO, LPP |
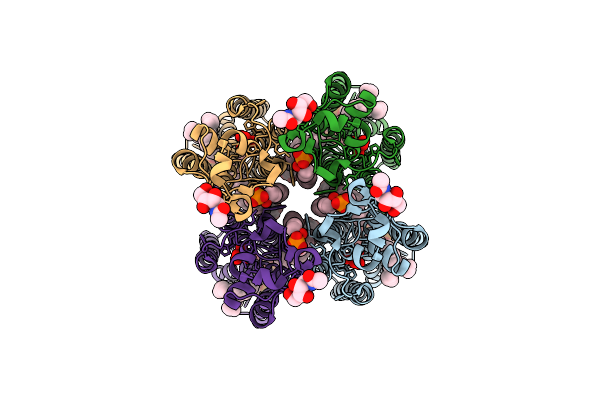 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: NAG, LBN, PT5, VO4, LPP |
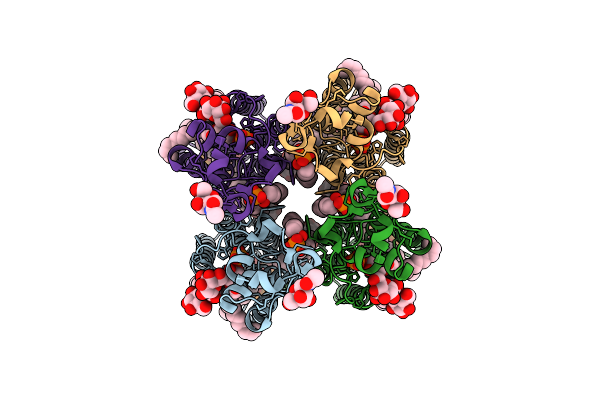 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: NAG, LBN, PT5, AV0, ZP7, PO4, LPP |
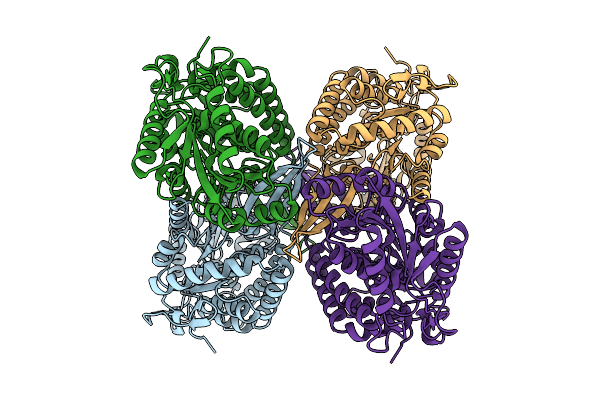 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Organism: Homo sapiens, Oplophorus gracilirostris, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: HOH |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: GSH, FBP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: PC1 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: VIRAL PROTEIN |
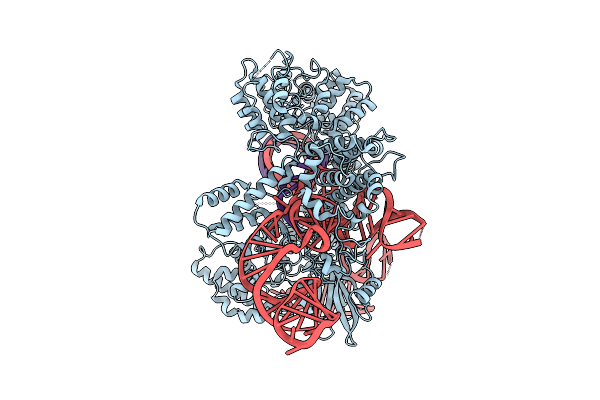 |
Cryo-Em Structure Of Frcas9 In Complex With Sgrna And 26-Nt Ts And 4-Nt Nts Substrates
Organism: Faecalibaculum rodentium, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE/RNA/DNA Ligands: HOH |
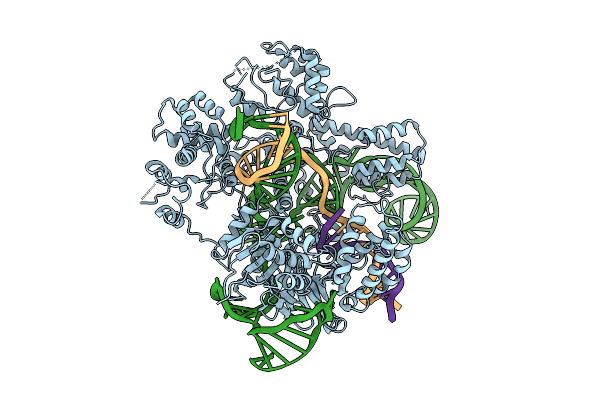 |
Cryo-Em Structure Of Frcas9 In Complex With Sgrna And 43-Bp Dsdna Substrate
Organism: Homo sapiens, Faecalibaculum rodentium
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA/RNA/HYDROLASE |
 |
Cryoem Structure Of Methylmalonic Acid Semialdehyde Dehydrogenase From Burkholderia Cenocepacia At 2.38A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: NAD |
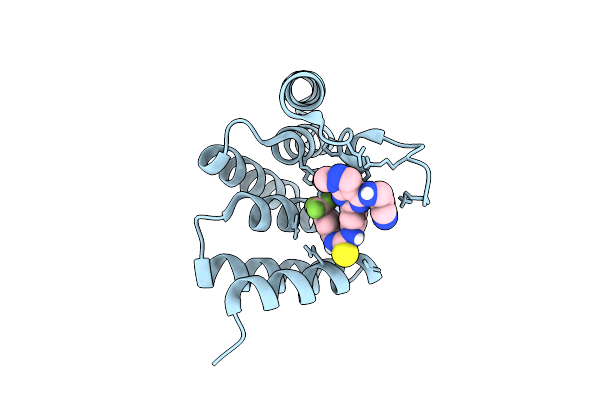 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: CELL CYCLE Ligands: A1JBJ |
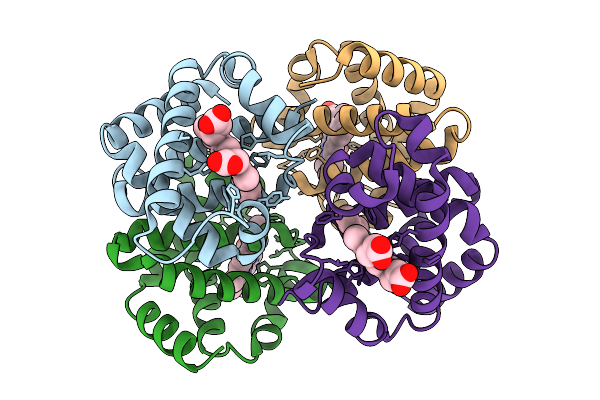 |
Work Experience Structure Of Bovine Hemoglobin, Collected At Room Temperature.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHT, CL, MG |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17B
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHV, IMD |

